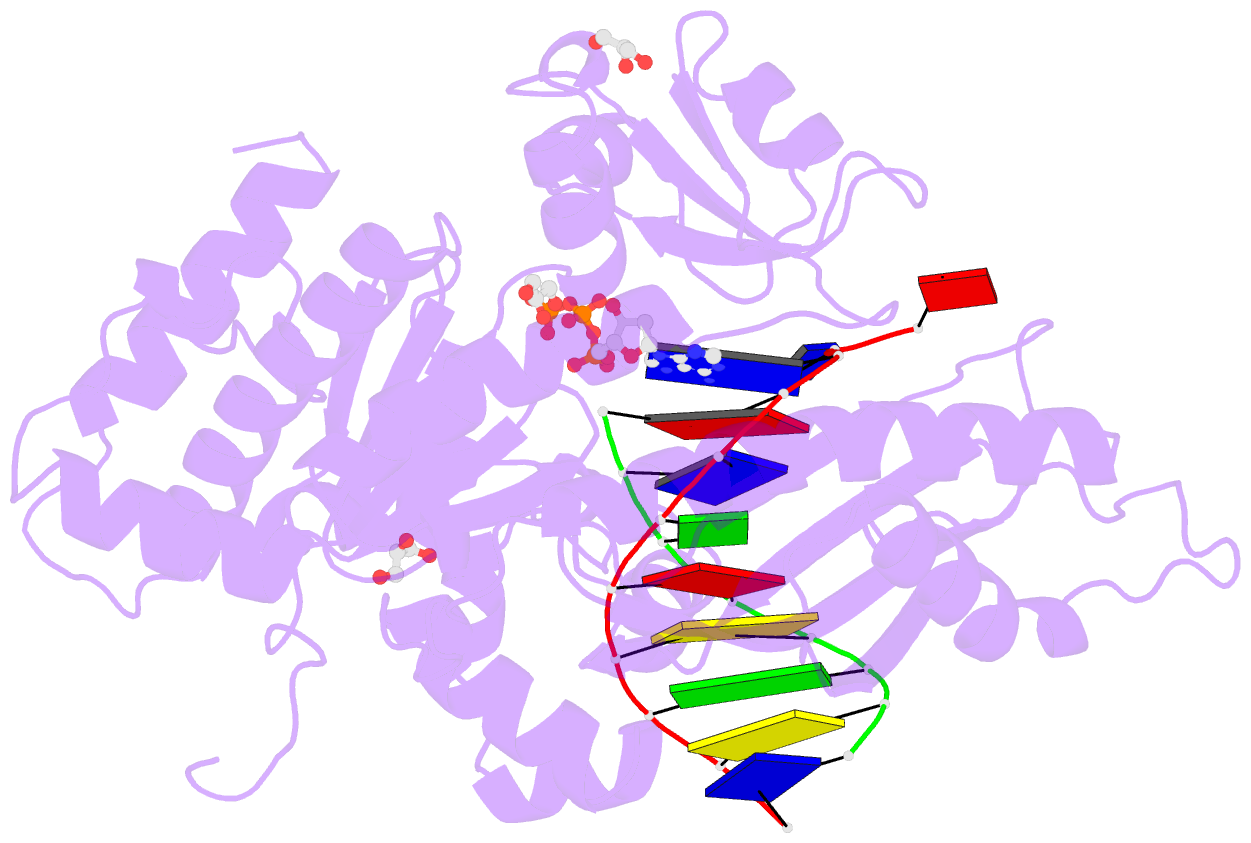
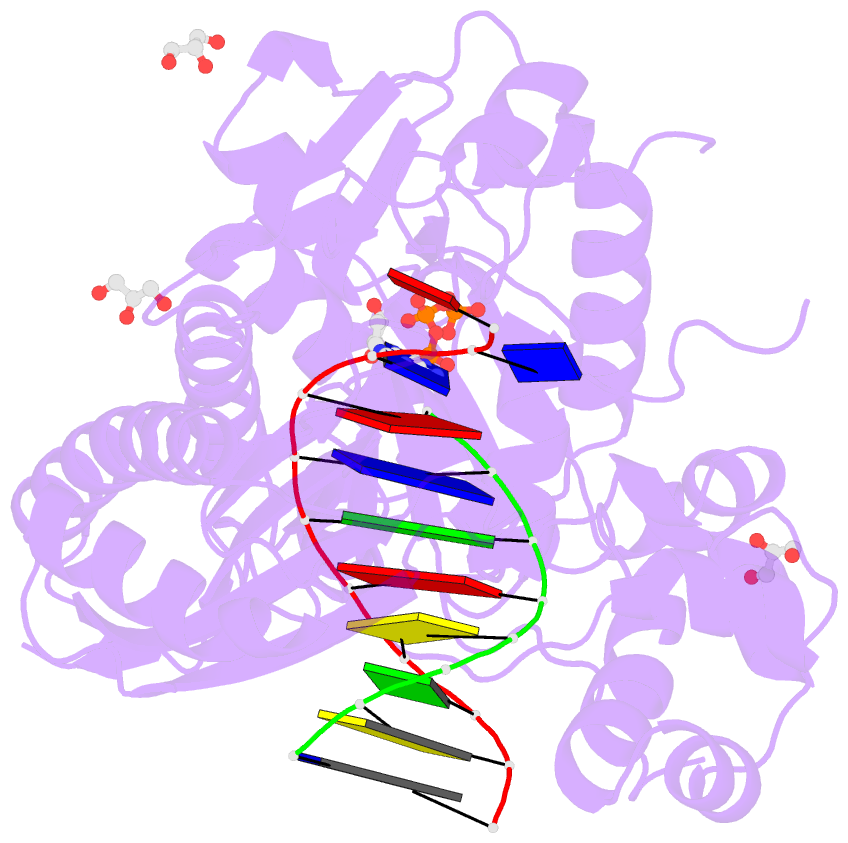
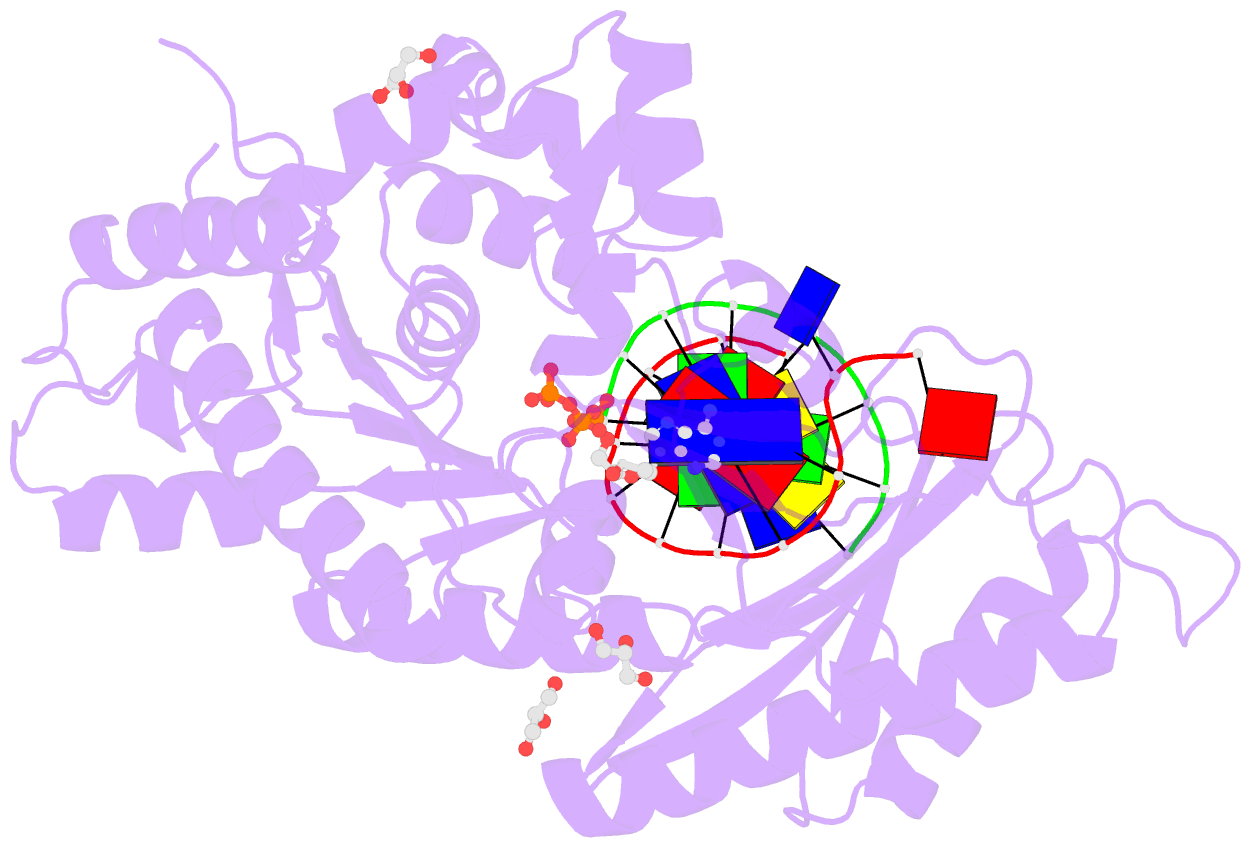
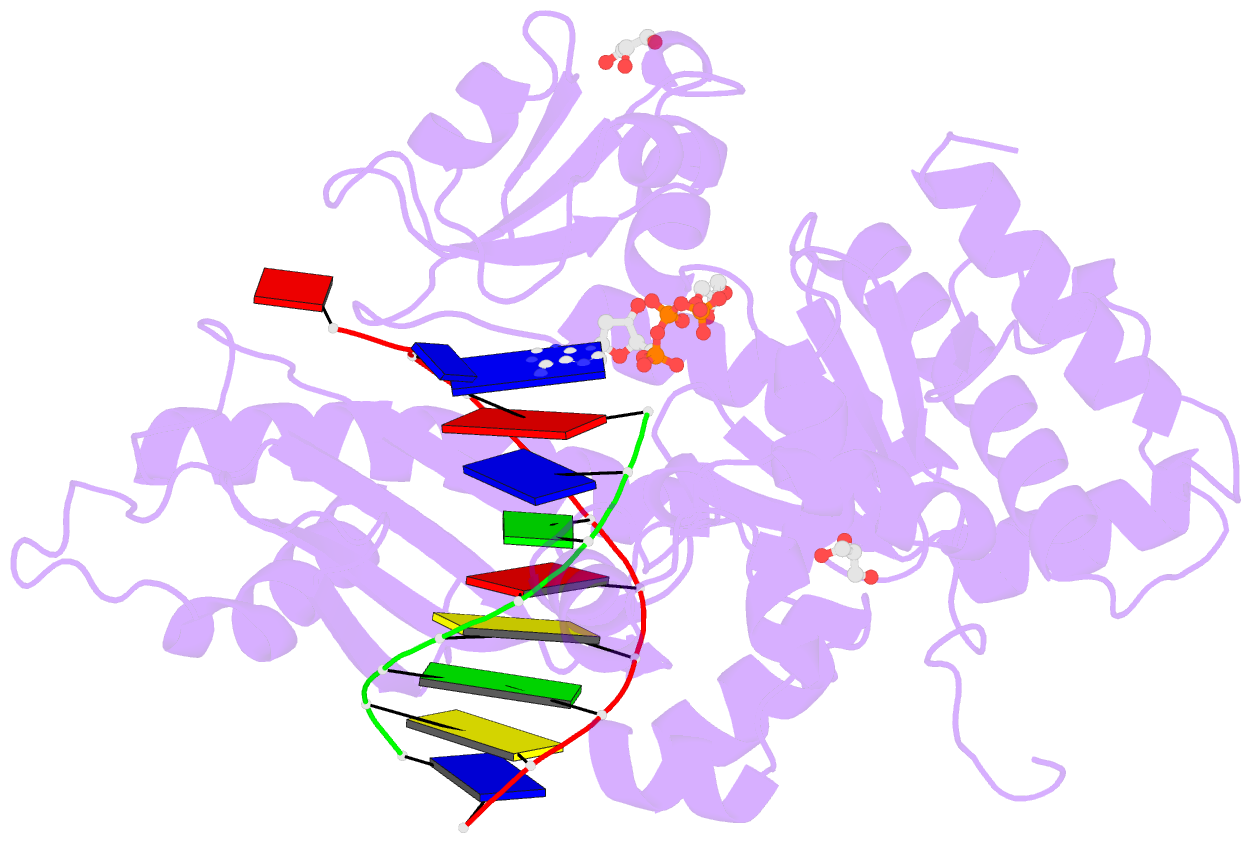
Summary information and primary citation
- PDB-id
- 7m7u; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (1.94 Å)
- Summary
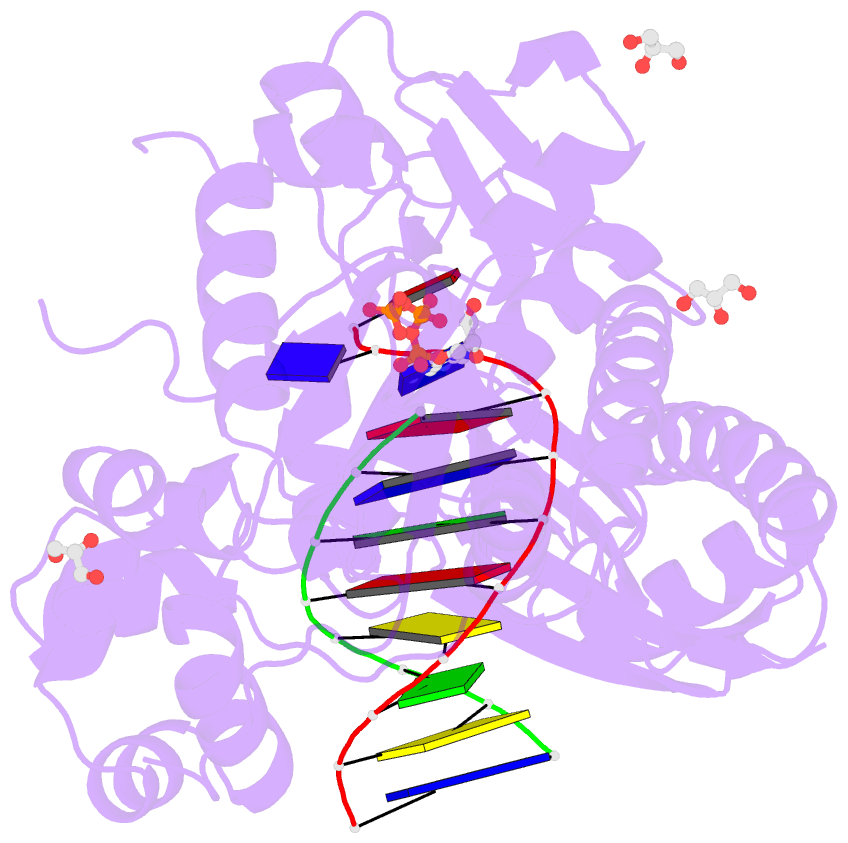
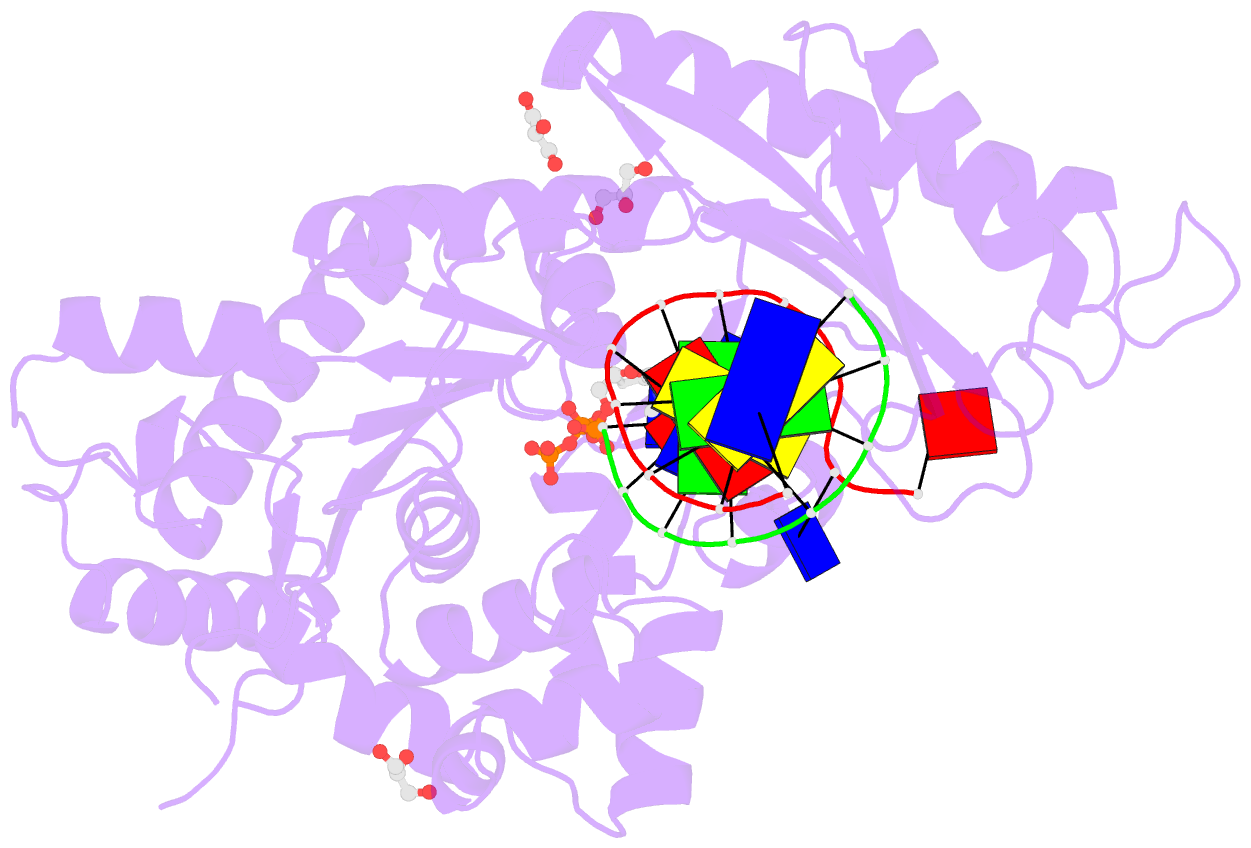
- Human DNA pol eta s113a with dt-ended primer and datp: in crystallo reaction for 480s
- Reference
- Gregory MT, Gao Y, Cui Q, Yang W (2021): "Multiple deprotonation paths of the nucleophile 3'-OH in the DNA synthesis reaction." Proc.Natl.Acad.Sci.USA, 118. doi: 10.1073/pnas.2103990118.
- Abstract
- DNA synthesis by polymerases is essential for life. Deprotonation of the nucleophile 3'-OH is thought to be the obligatory first step in the DNA synthesis reaction. We have examined each entity surrounding the nucleophile 3'-OH in the reaction catalyzed by human DNA polymerase (Pol) η and delineated the deprotonation process by combining mutagenesis with steady-state kinetics, high-resolution structures of in crystallo reactions, and molecular dynamics simulations. The conserved S113 residue, which forms a hydrogen bond with the primer 3'-OH in the ground state, stabilizes the primer end in the active site. Mutation of S113 to alanine destabilizes primer binding and reduces the catalytic efficiency. Displacement of a water molecule that is hydrogen bonded to the 3'-OH using the 2'-OH of a ribonucleotide or 2'-F has little effect on catalysis. Moreover, combining the S113A mutation with 2'-F replacement, which removes two potential hydrogen acceptors of the 3'-OH, does not reduce the catalytic efficiency. We conclude that the proton can leave the O3' via alternative paths, supporting the hypothesis that binding of the third Mg2+ initiates the reaction by breaking the α-β phosphodiester bond of an incoming deoxyribonucleoside triphosphate (dNTP).