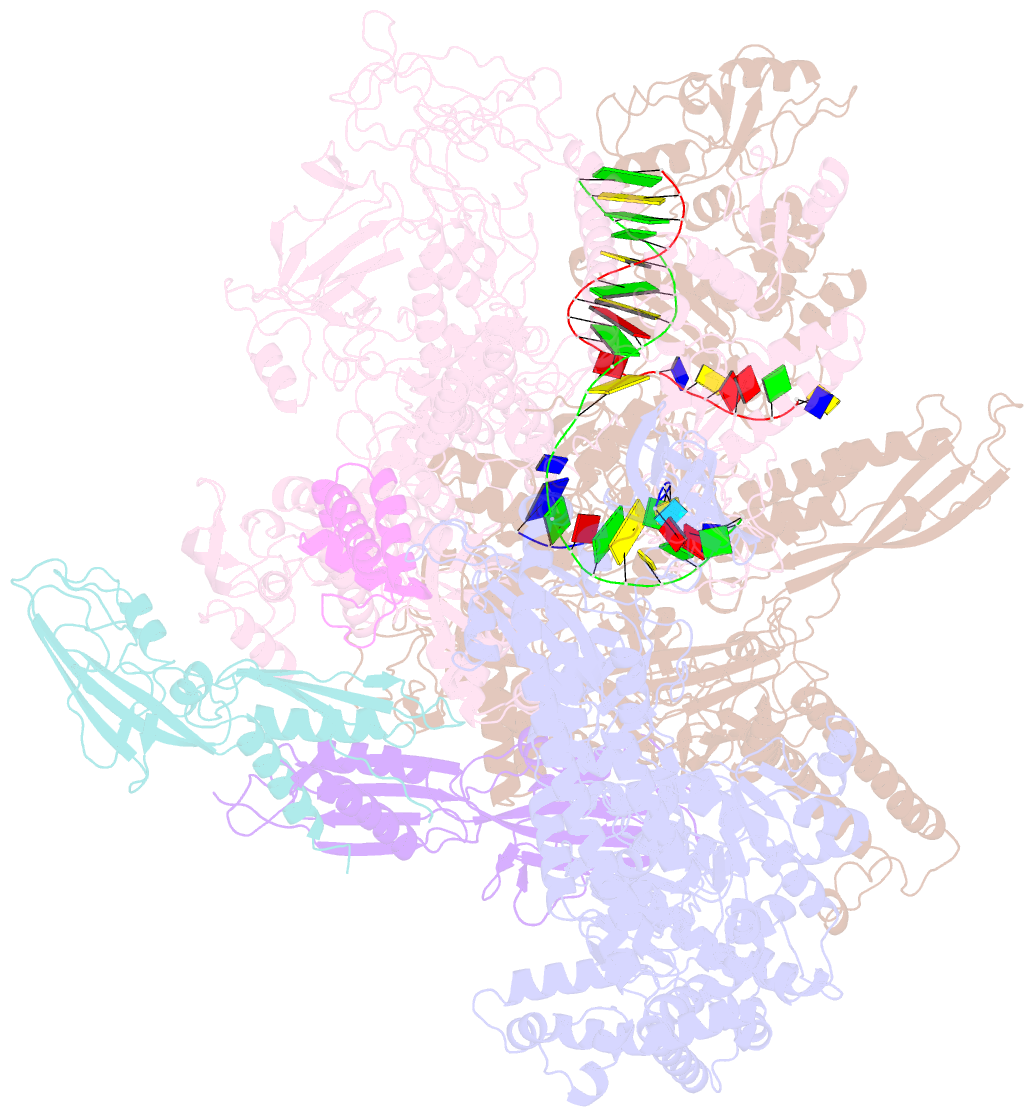
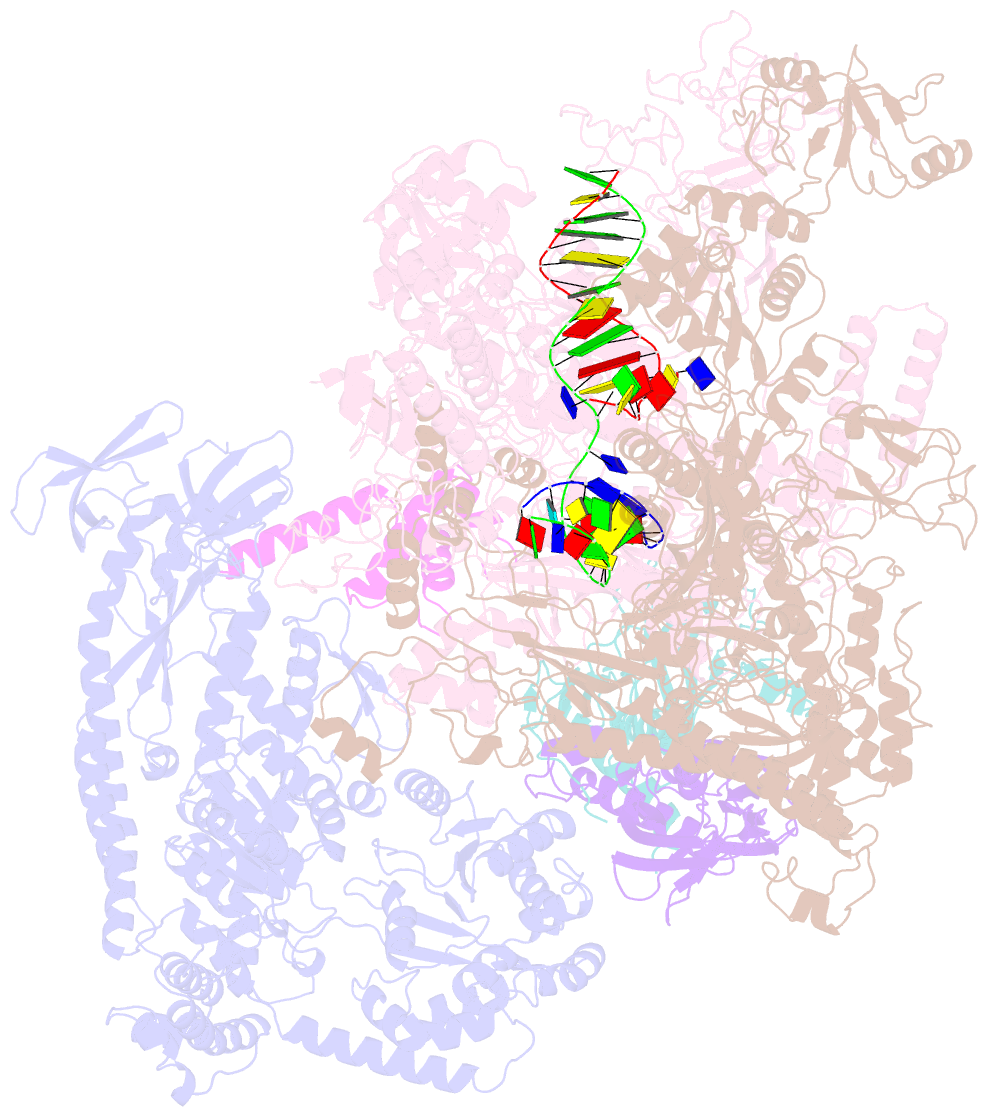
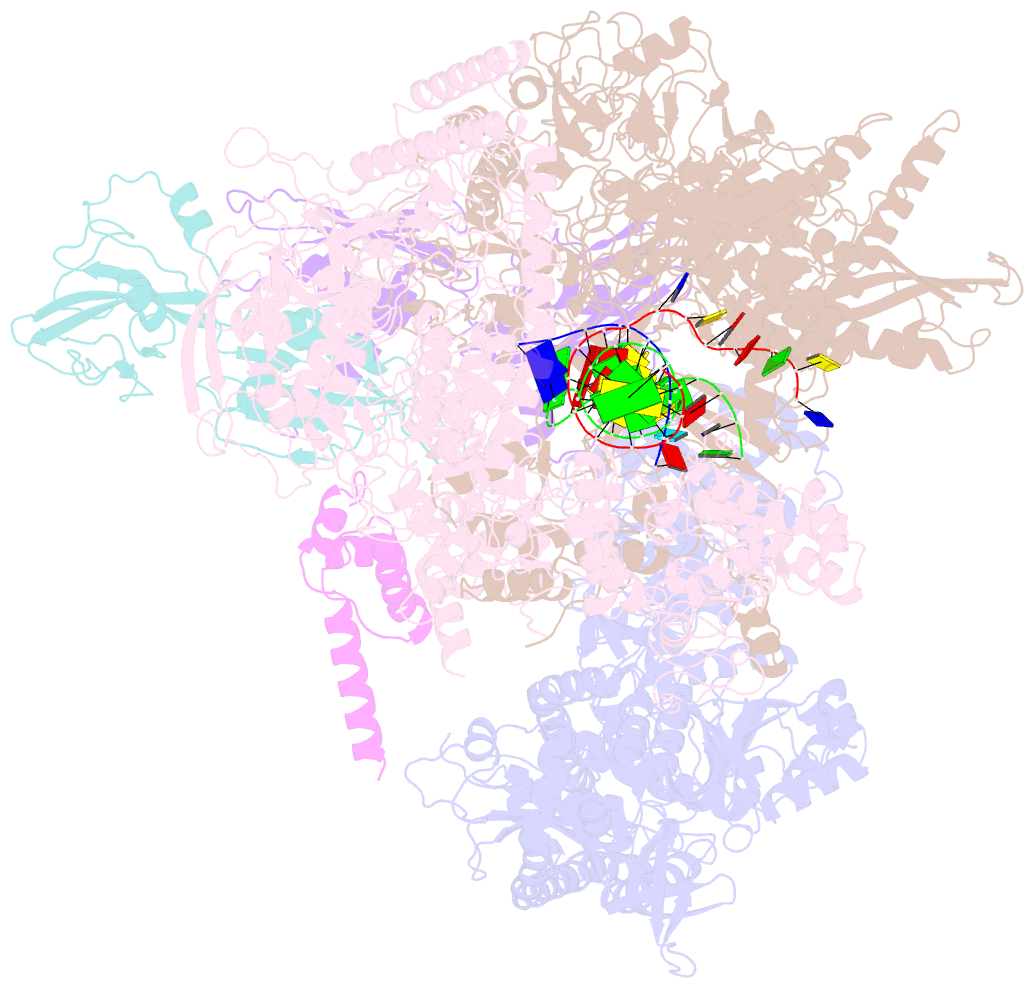
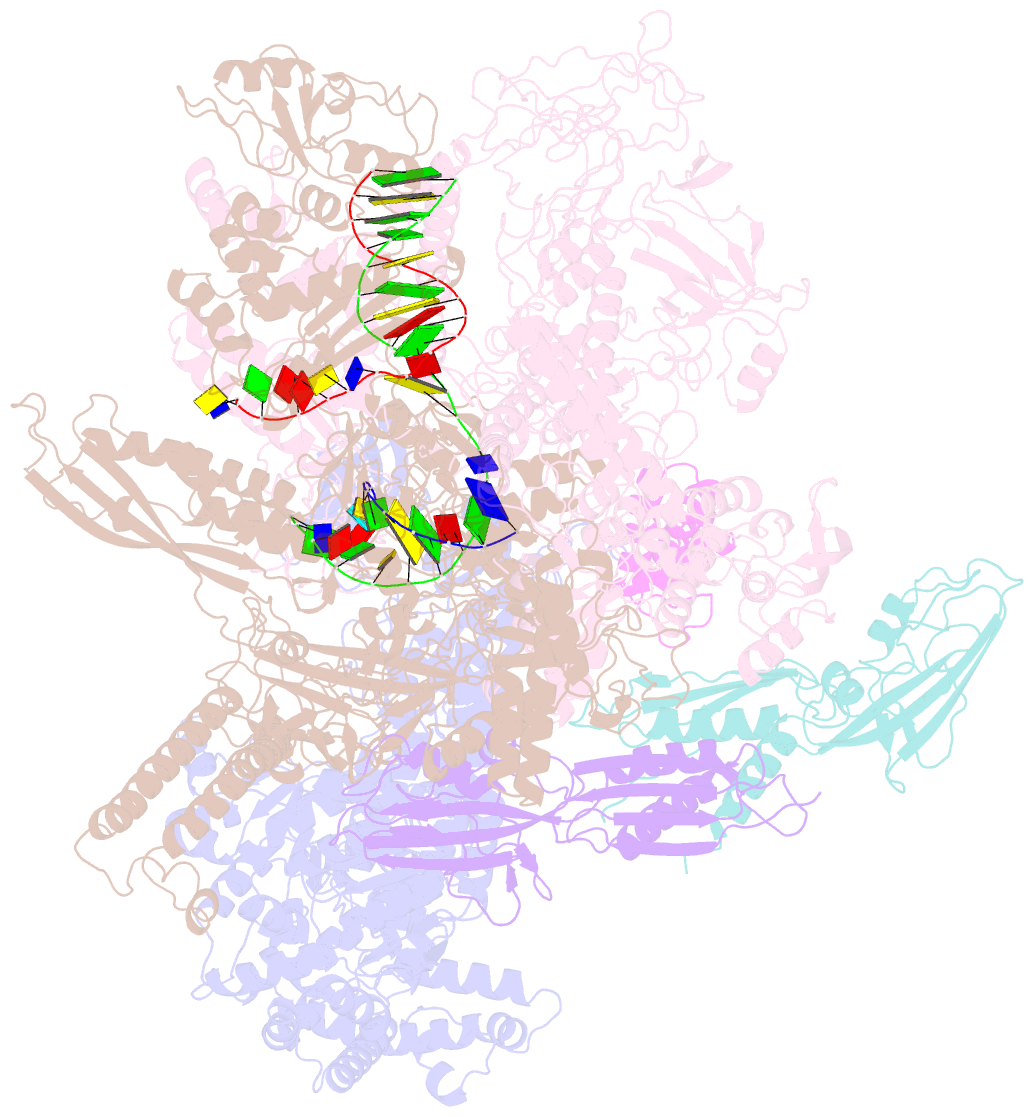
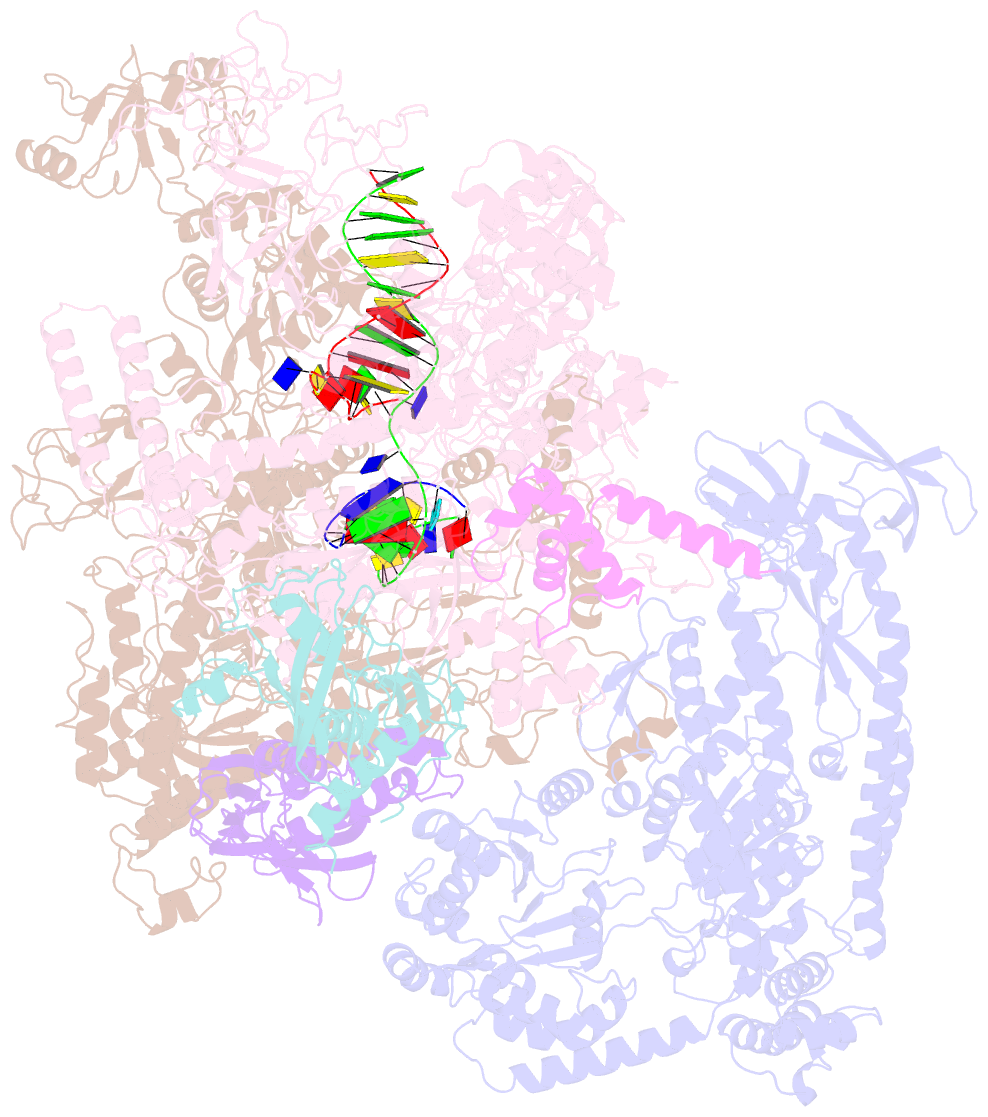
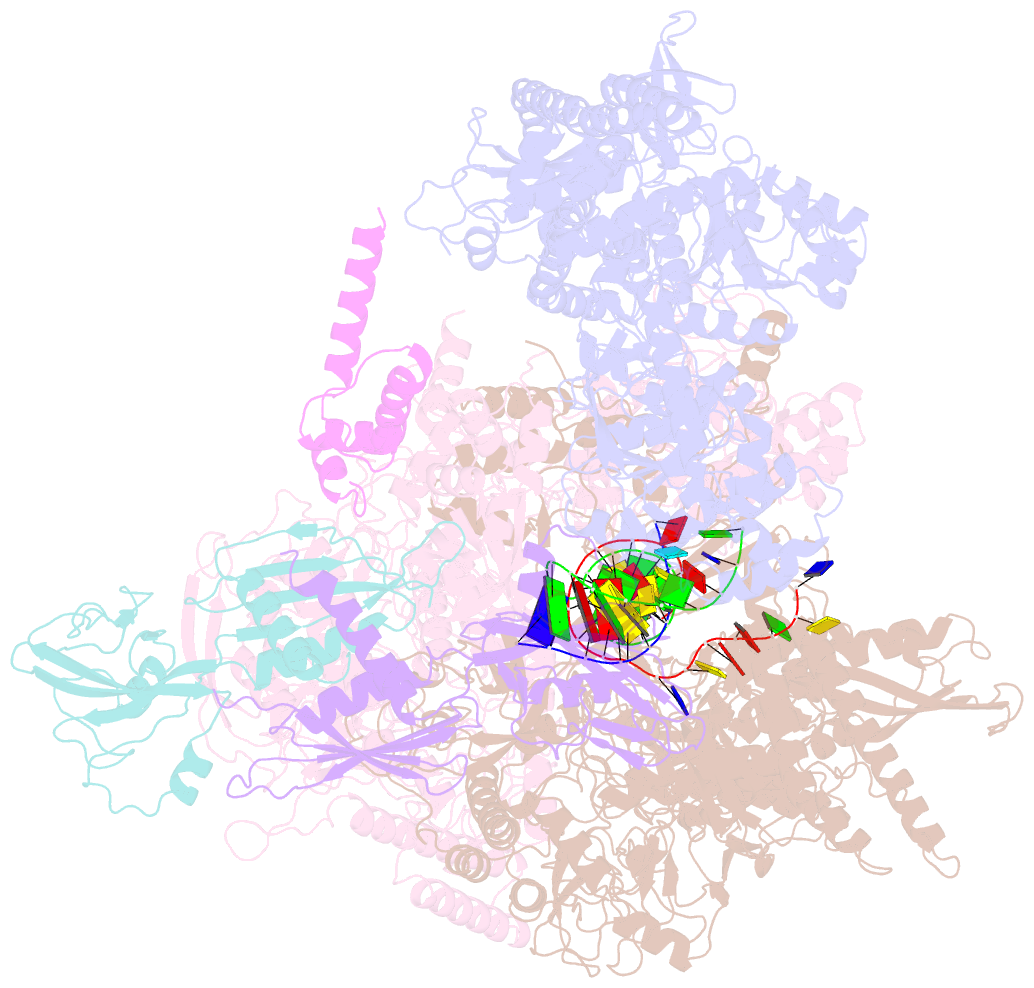
Summary information and primary citation
- PDB-id
- 7m8e; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-hydrolase-DNA-RNA
- Method
- cryo-EM (3.4 Å)
- Summary
- E.coli rnap-rapa elongation complex
- Reference
- Shi W, Zhou W, Chen M, Yang Y, Hu Y, Liu B (2021): "Structural basis for activation of Swi2/Snf2 ATPase RapA by RNA polymerase." Nucleic Acids Res., 49, 10707-10716. doi: 10.1093/nar/gkab744.
- Abstract
- RapA is a bacterial RNA polymerase (RNAP)-associated Swi2/Snf2 ATPase that stimulates RNAP recycling. The ATPase activity of RapA is autoinhibited by its N-terminal domain (NTD) but activated with RNAP bound. Here, we report a 3.4-Å cryo-EM structure of Escherichia coli RapA-RNAP elongation complex, in which the ATPase active site of RapA is structurally remodeled. In this process, the NTD of RapA is wedged open by RNAP β' zinc-binding domain (ZBD). In addition, RNAP β flap tip helix (FTH) forms extensive hydrophobic interactions with RapA ATPase core domains. Functional assay demonstrates that removing the ZBD or FTH of RNAP significantly impairs its ability to activate the ATPase activity of RapA. Our results provide the structural basis of RapA ATPase activation by RNAP, through the active site remodeling driven by the ZBD-buttressed large-scale opening of NTD and the direct interactions between FTH and ATPase core domains.