Summary information and primary citation
- PDB-id
- 7mbn; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA binding protein-DNA
- Method
- cryo-EM
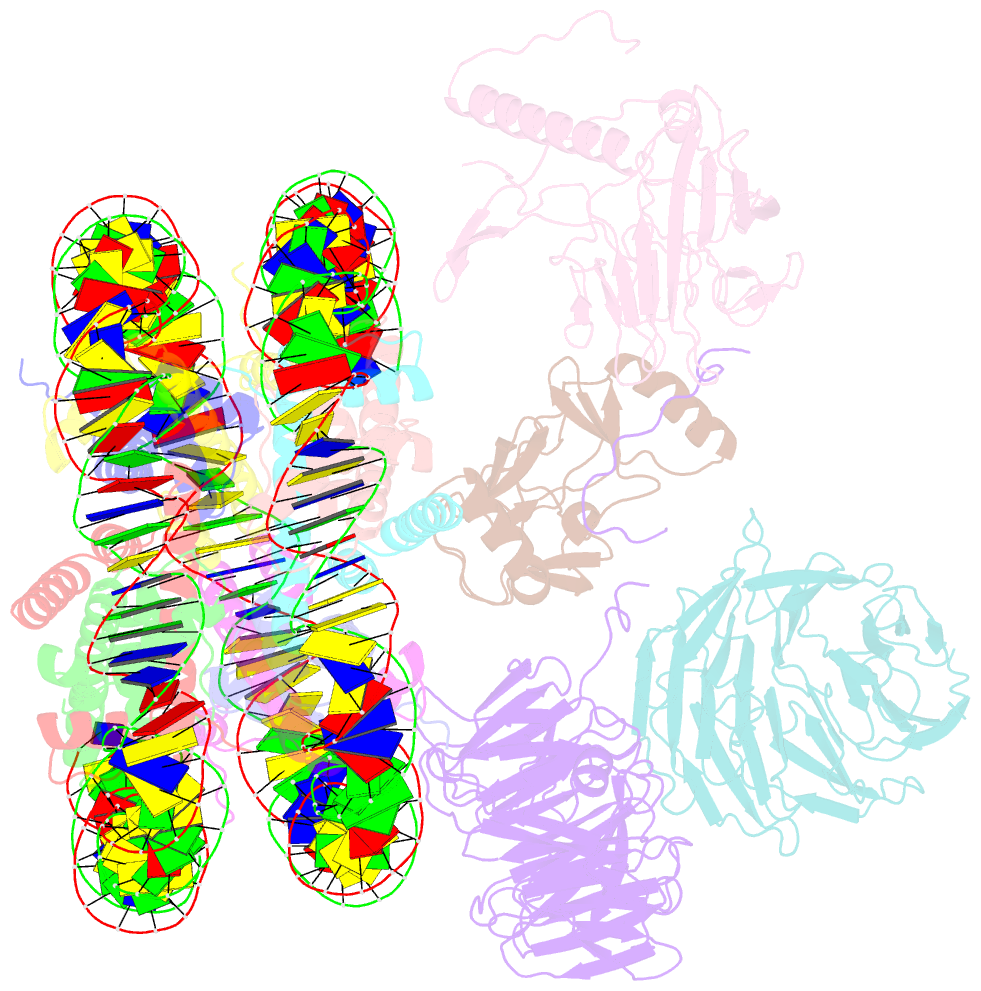
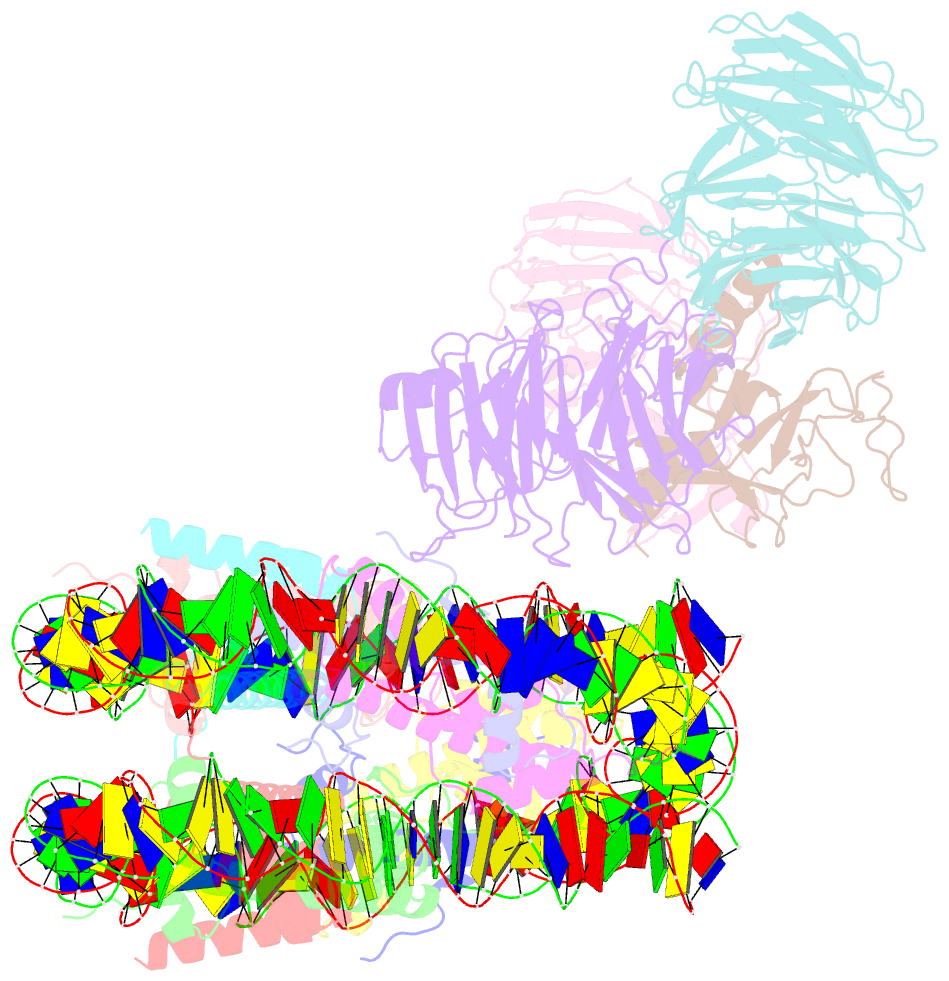
- Summary
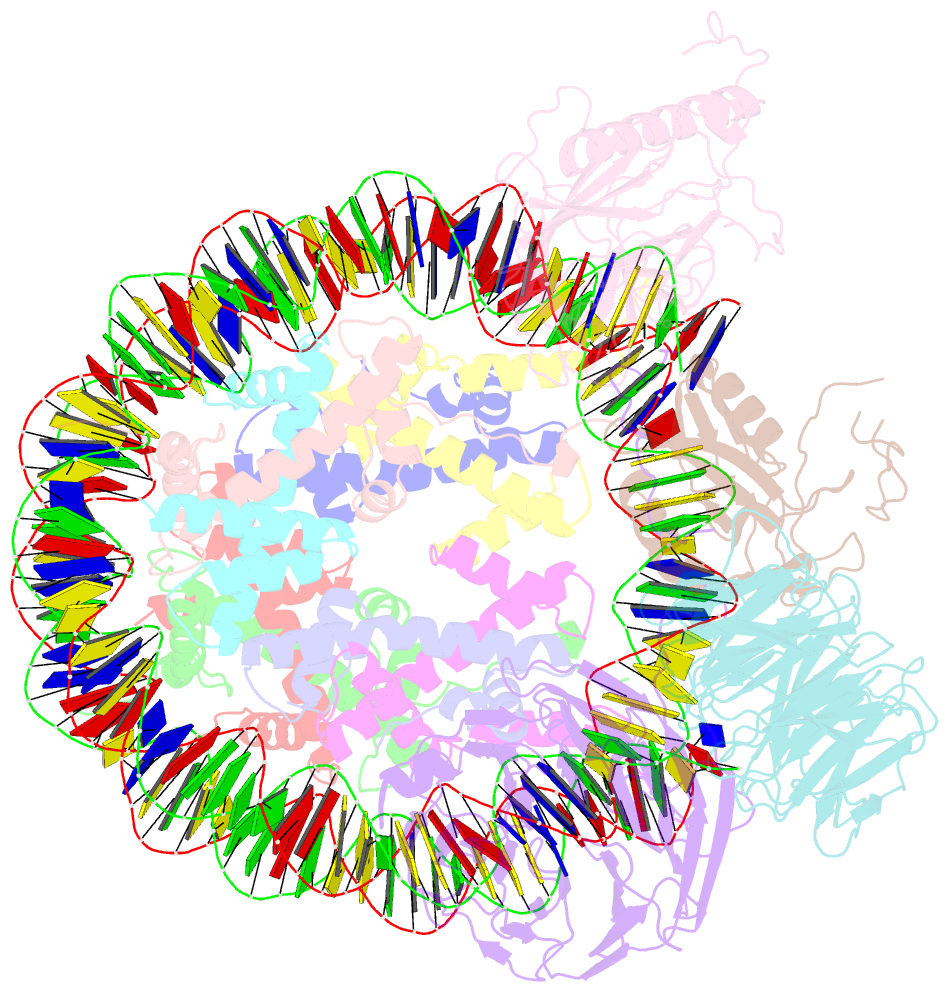
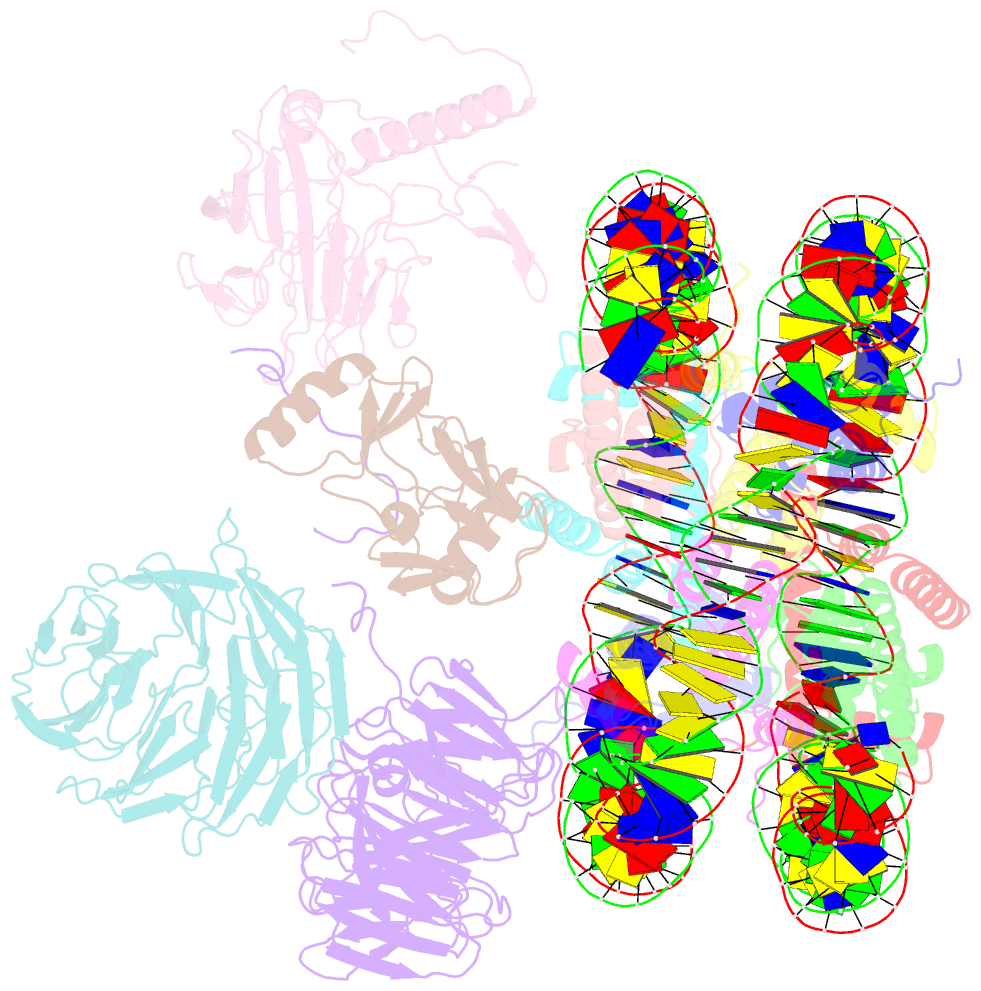
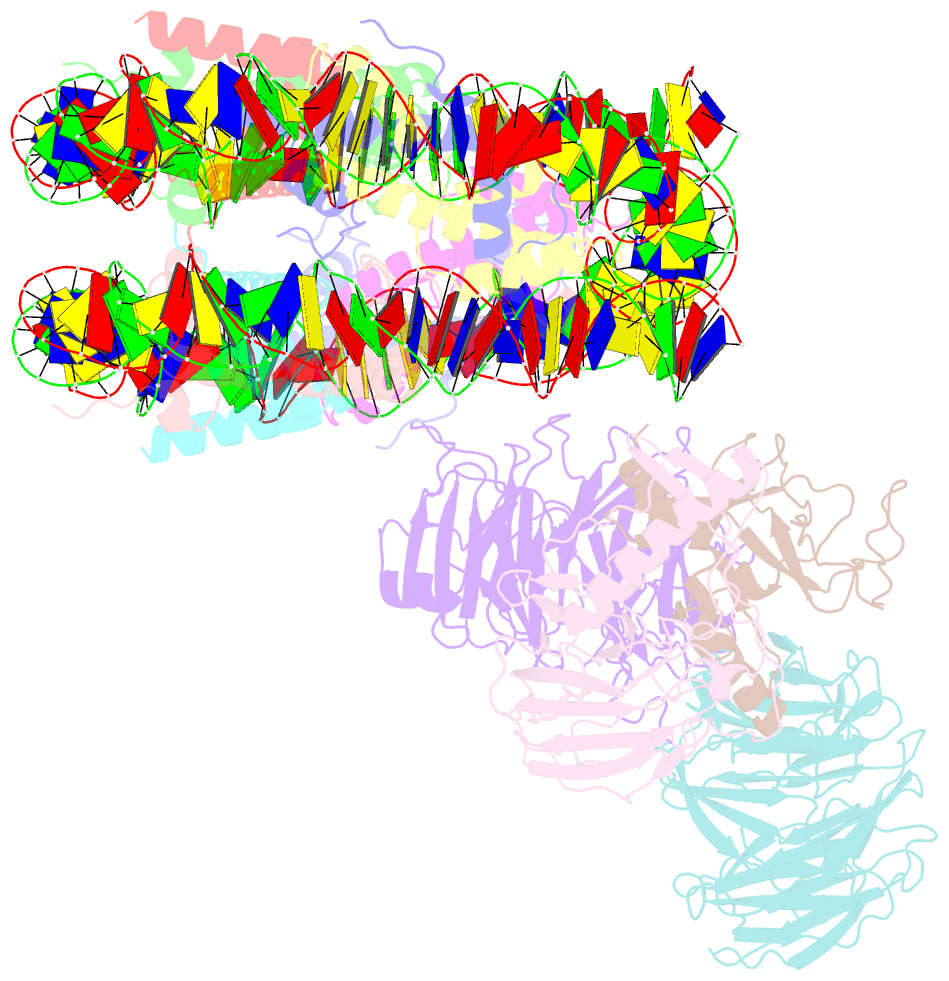
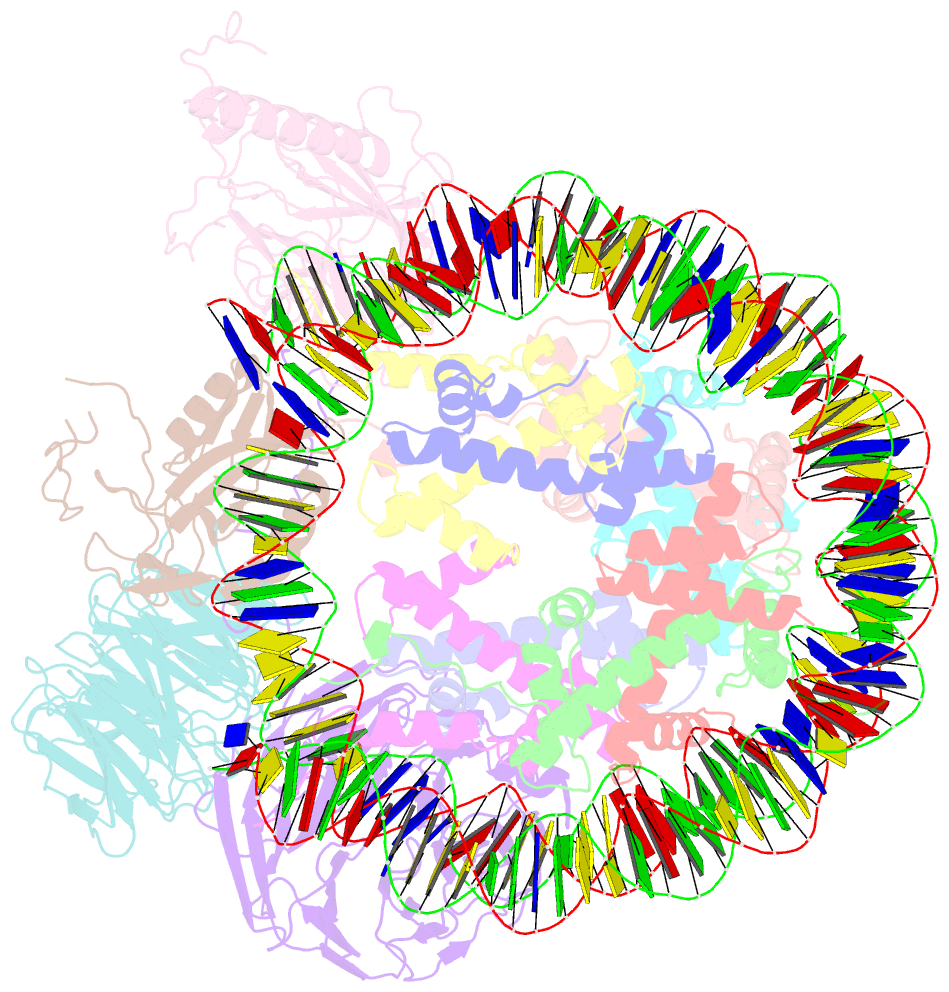
- cryo-EM structure of mll1-ncp (h3k4m) complex, mode02
- Reference
- Ayoub A, Park SH, Lee YT, Cho US, Dou Y (2022): "Regulation of MLL1 Methyltransferase Activity in Two Distinct Nucleosome Binding Modes." Biochemistry, 61, 1-9. doi: 10.1021/acs.biochem.1c00603.
- Abstract
- Cryo-EM structures of the KMT2A/MLL1 core complex bound on nucleosome core particles (NCPs) suggest unusual rotational dynamics of the MLL1 complex approaching its physiological substrate. However, the functional implication of such dynamics remains unclear. Here, we show that the MLL1 core complex also shows high rotational dynamics bound on the NCP carrying the catalytically inert histone H3 lysine 4 to methionine (K4M) mutation. There are two major binding modes of the MLL1 complex on the NCPK4M. Importantly, disruption of only one of the binding modes compromised the overall MLL1 activity in an NCP-specific manner. We propose that the MLL1 core complex probably exists in an equilibrium of poised and active binding modes. The high rotational dynamics of the MLL1 complex on the NCP is a feature that can be exploited for loci-specific regulation of H3K4 methylation in higher eukaryotes.