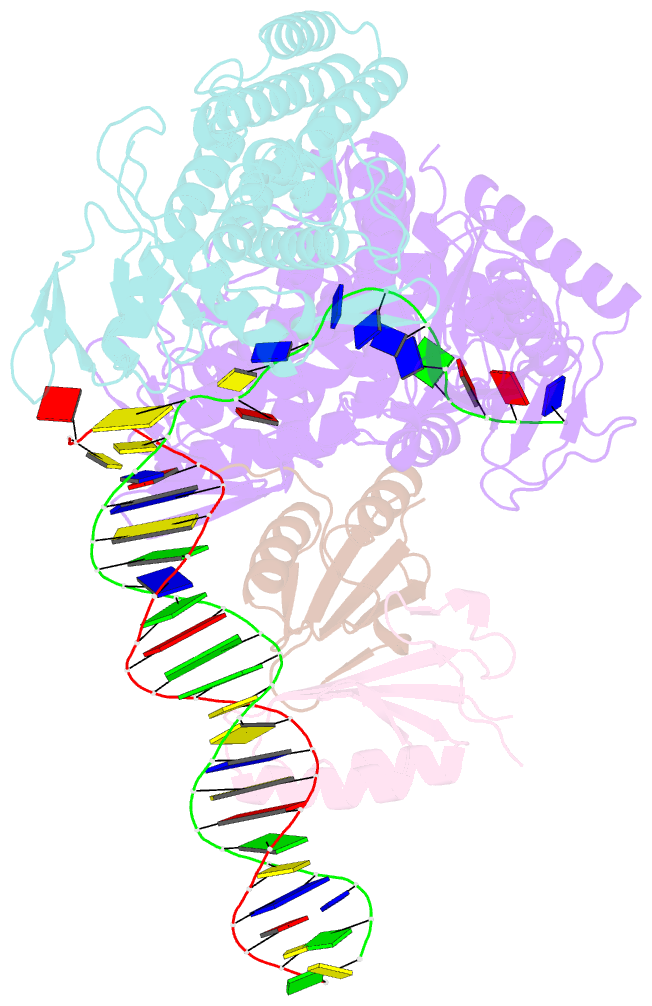
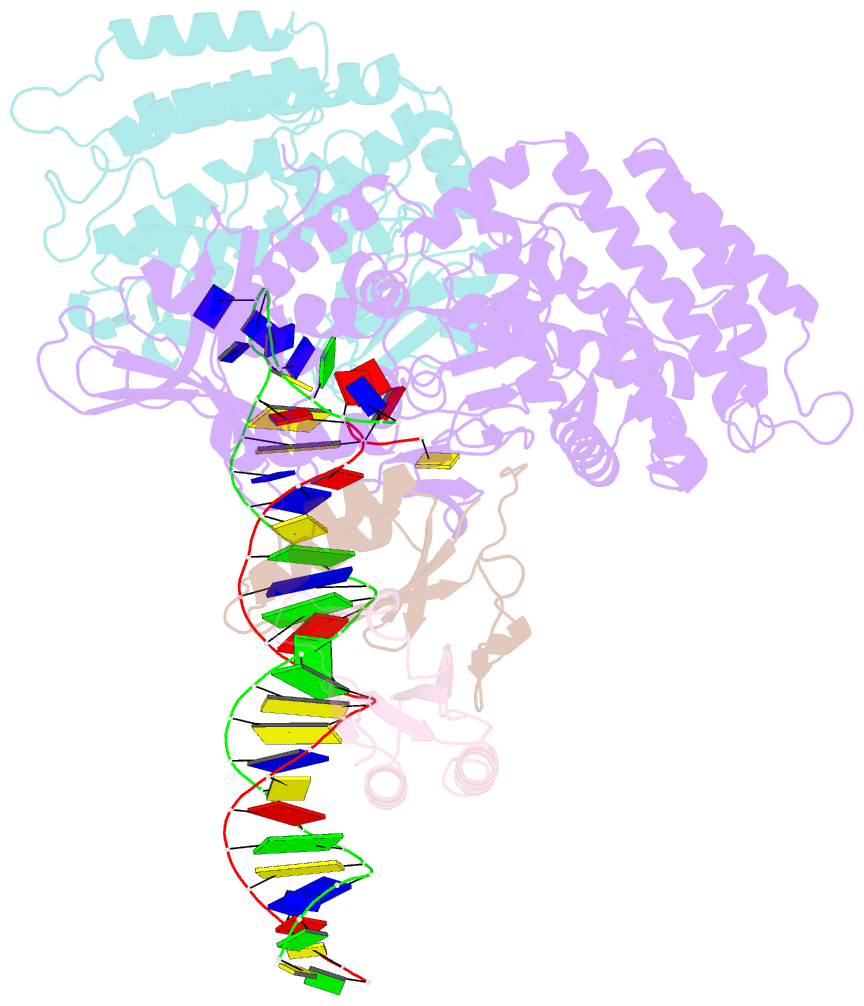
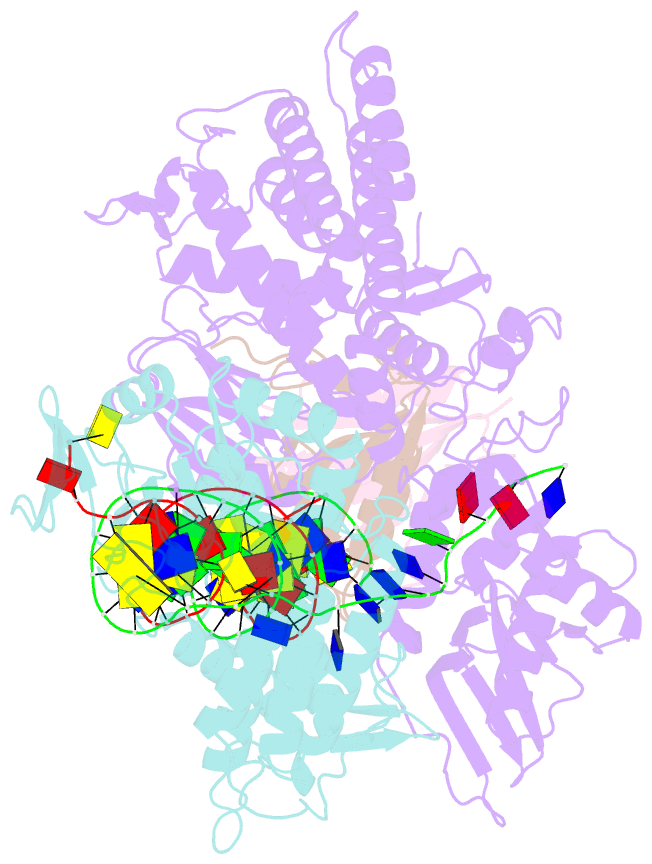
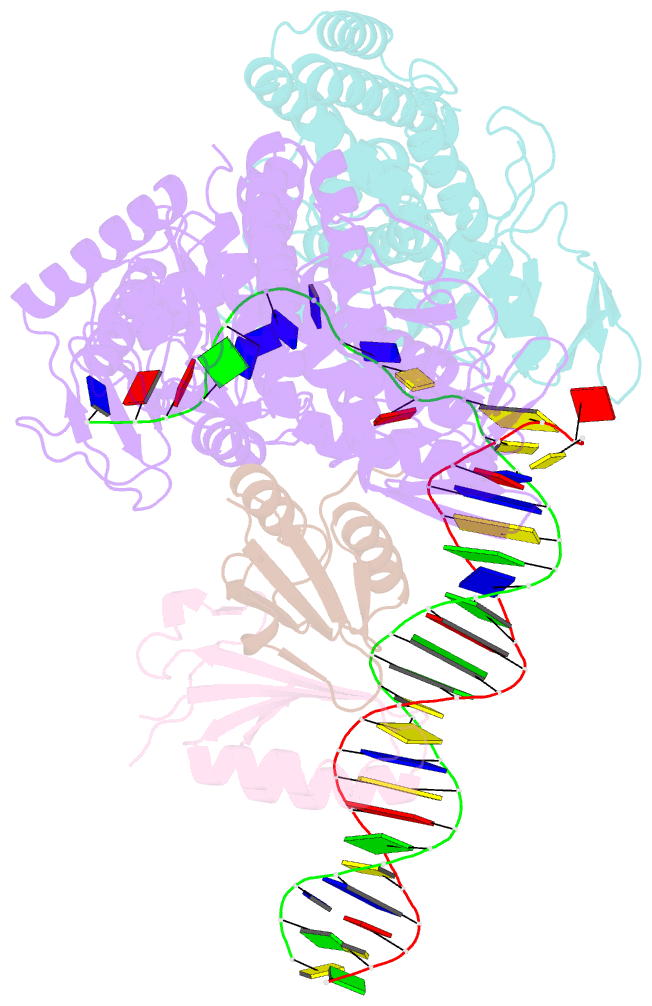
Summary information and primary citation
- PDB-id
- 7mid; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- cryo-EM (3.56 Å)
- Summary
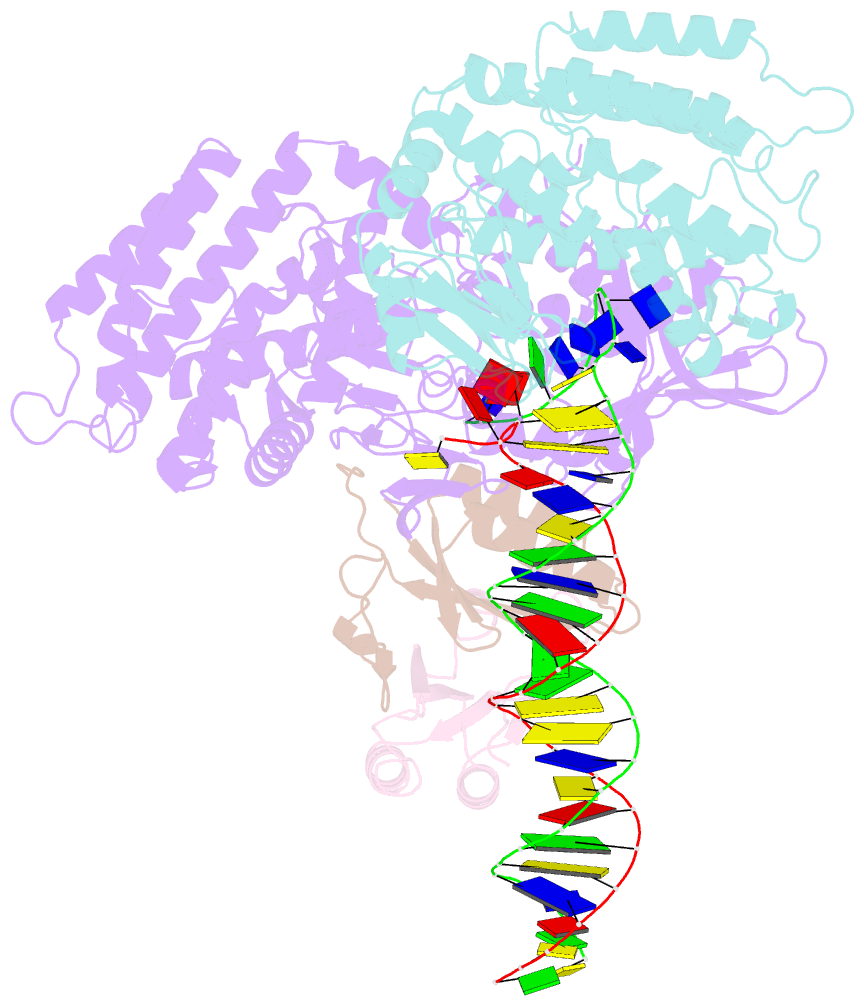
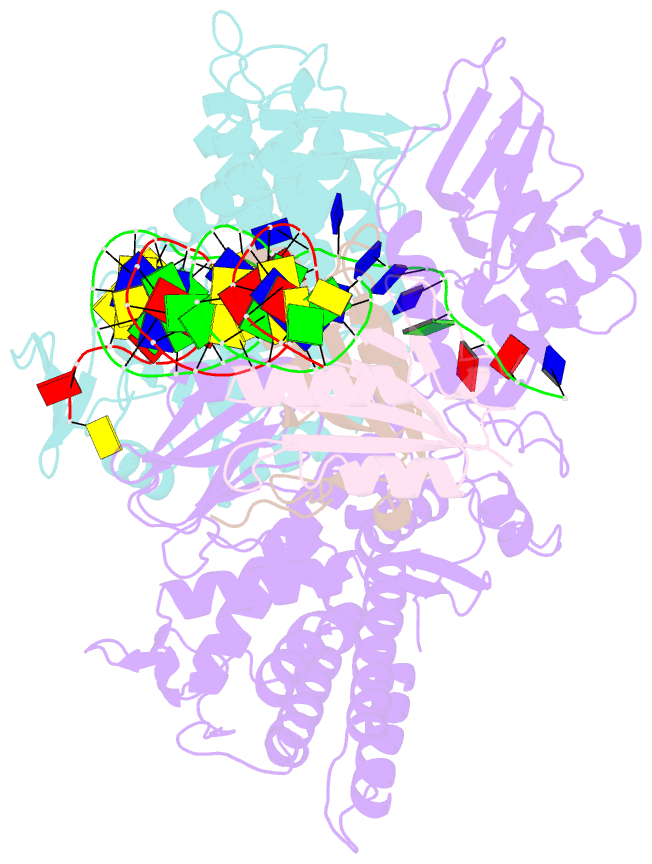
- Sub-complex of cas4-cas1-cas2 bound pam containing DNA
- Reference
- Hu C, Almendros C, Nam KH, Costa AR, Vink JNA, Haagsma AC, Bagde SR, Brouns SJJ, Ke A (2021): "Mechanism for Cas4-assisted directional spacer acquisition in CRISPR-Cas." Nature, 598, 515-520. doi: 10.1038/s41586-021-03951-z.
- Abstract
- Prokaryotes adapt to challenges from mobile genetic elements by integrating spacers derived from foreign DNA in the CRISPR array1. Spacer insertion is carried out by the Cas1-Cas2 integrase complex2-4. A substantial fraction of CRISPR-Cas systems use a Fe-S cluster containing Cas4 nuclease to ensure that spacers are acquired from DNA flanked by a protospacer adjacent motif (PAM)5,6 and inserted into the CRISPR array unidirectionally, so that the transcribed CRISPR RNA can guide target searching in a PAM-dependent manner. Here we provide a high-resolution mechanistic explanation for the Cas4-assisted PAM selection, spacer biogenesis and directional integration by type I-G CRISPR in Geobacter sulfurreducens, in which Cas4 is naturally fused with Cas1, forming Cas4/Cas1. During biogenesis, only DNA duplexes possessing a PAM-embedded 3'-overhang trigger Cas4/Cas1-Cas2 assembly. During this process, the PAM overhang is specifically recognized and sequestered, but is not cleaved by Cas4. This 'molecular constipation' prevents the PAM-side prespacer from participating in integration. Lacking such sequestration, the non-PAM overhang is trimmed by host nucleases and integrated to the leader-side CRISPR repeat. Half-integration subsequently triggers PAM cleavage and Cas4 dissociation, allowing spacer-side integration. Overall, the intricate molecular interaction between Cas4 and Cas1-Cas2 selects PAM-containing prespacers for integration and couples the timing of PAM processing with the stepwise integration to establish directionality.