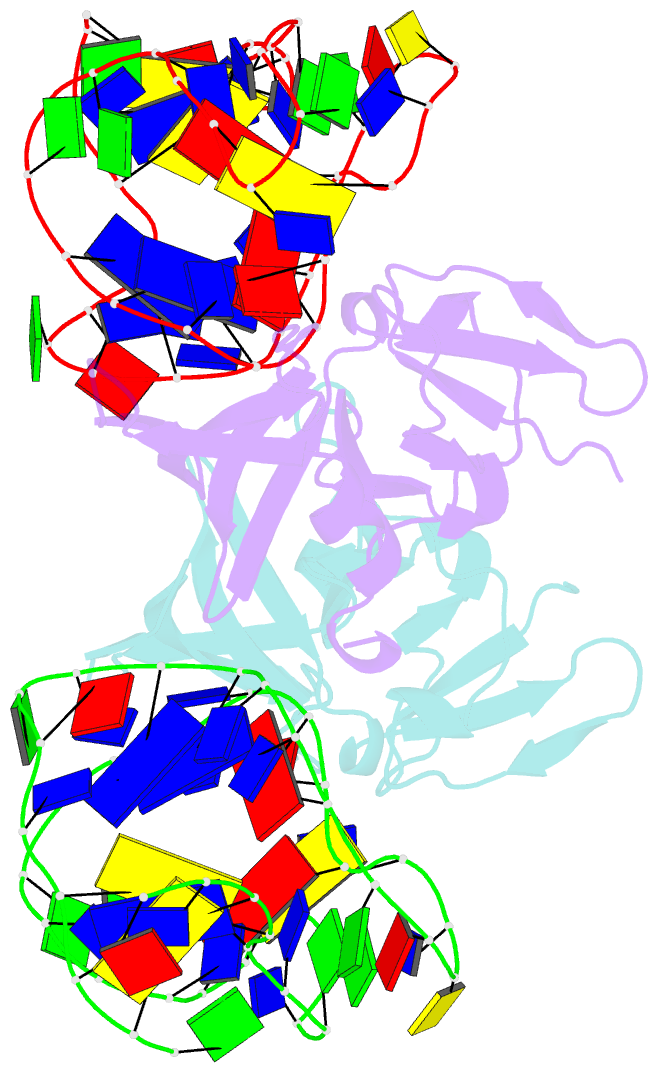
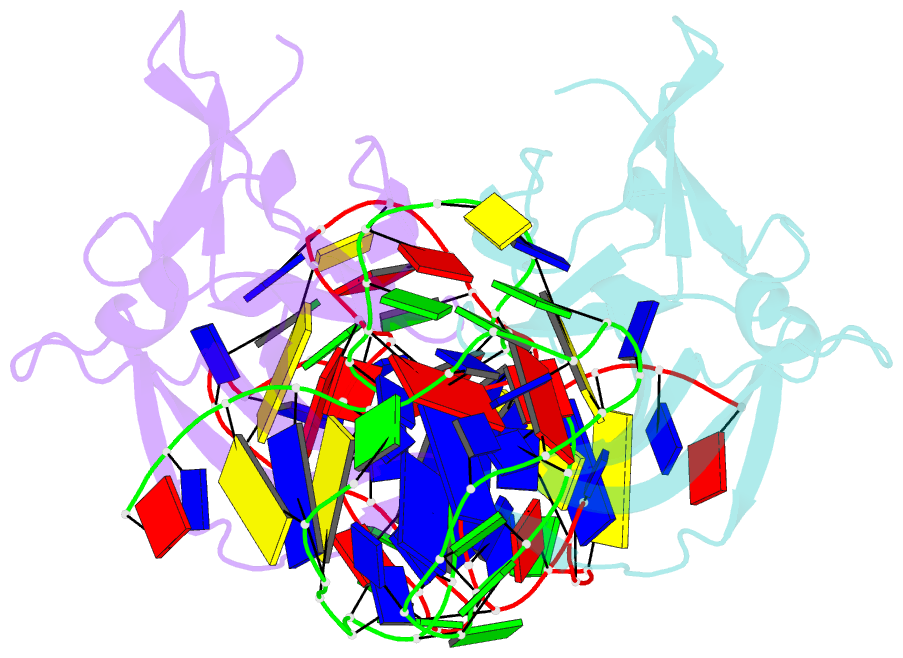
Summary information and primary citation
- PDB-id
- 7mk1; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- immune system-DNA
- Method
- X-ray (1.9 Å)
- Summary
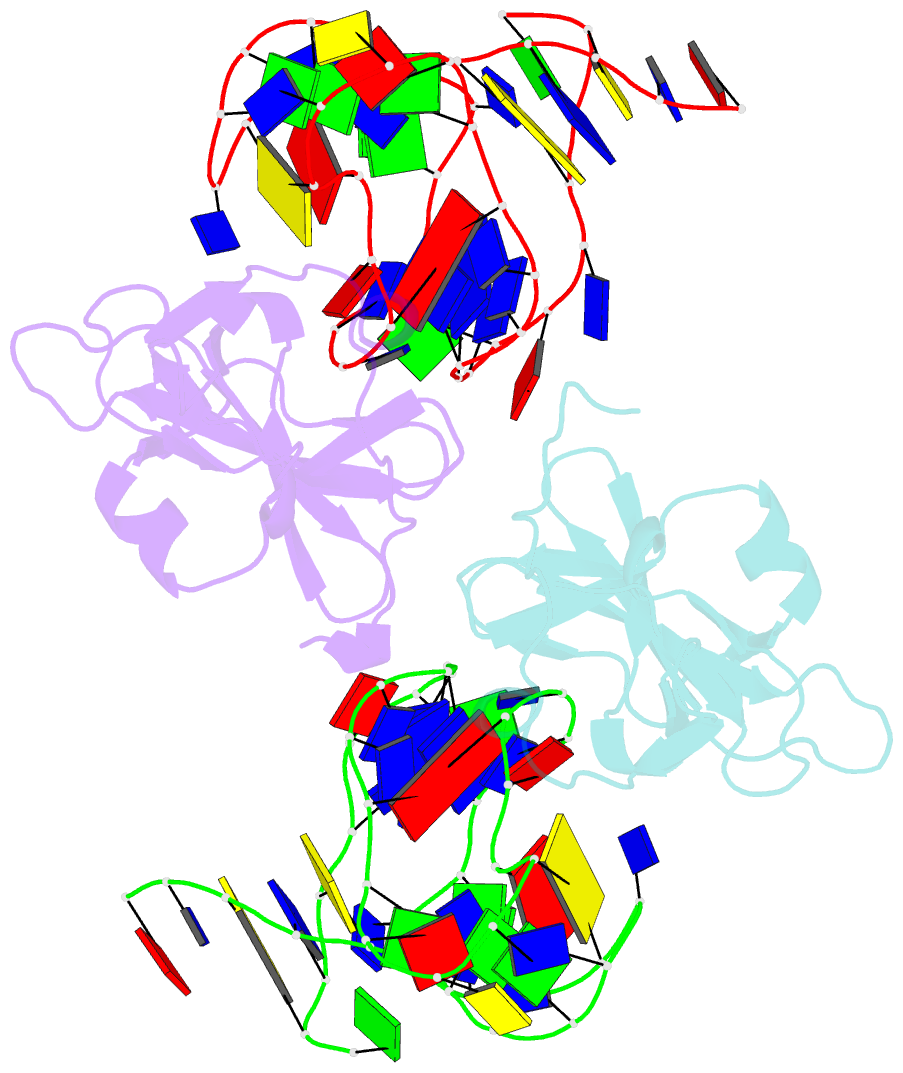
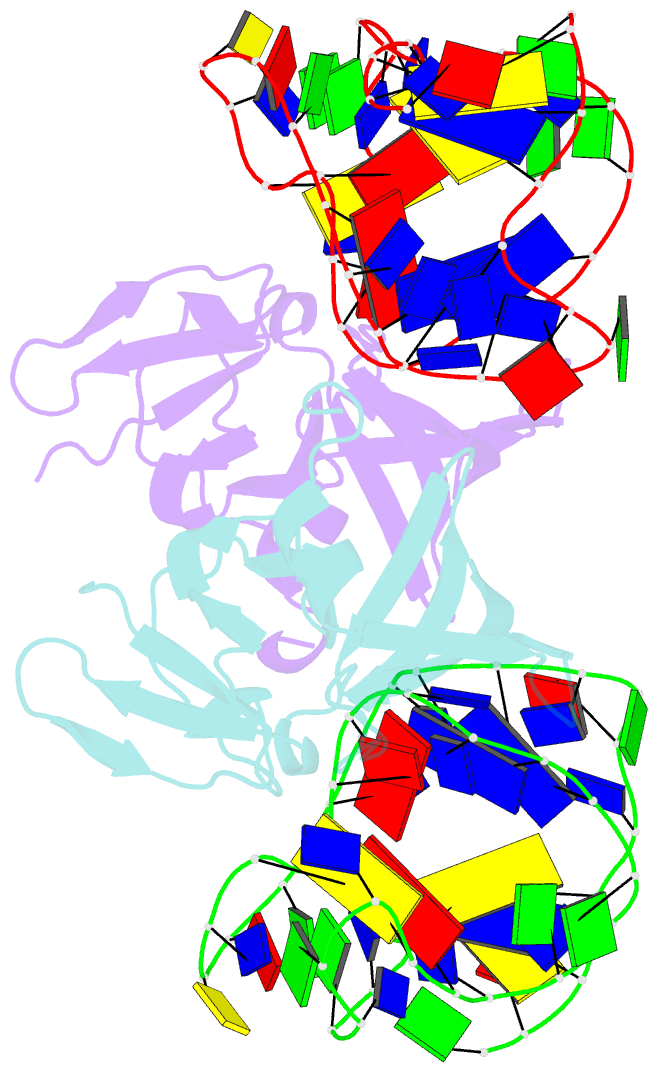
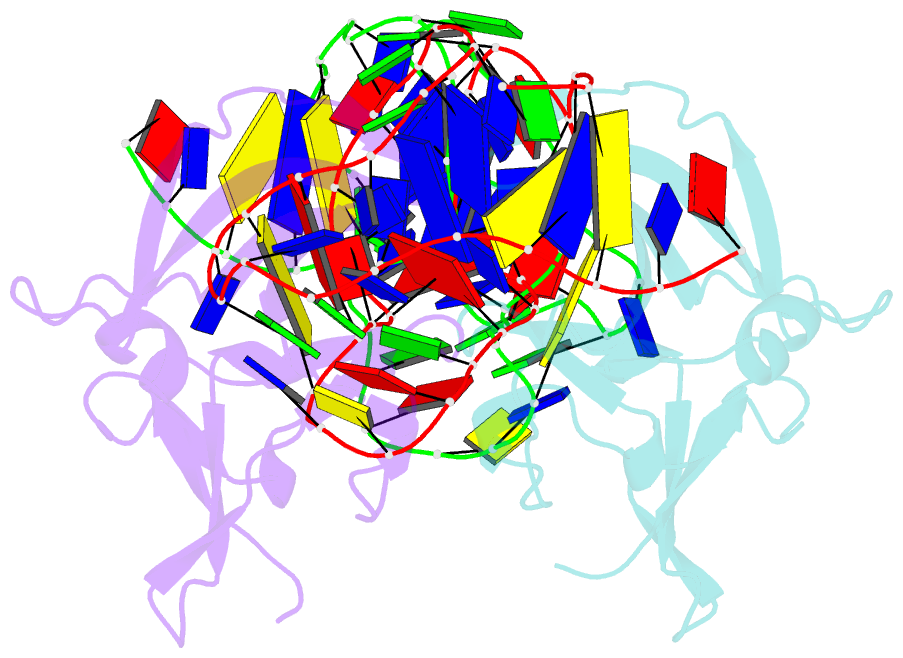
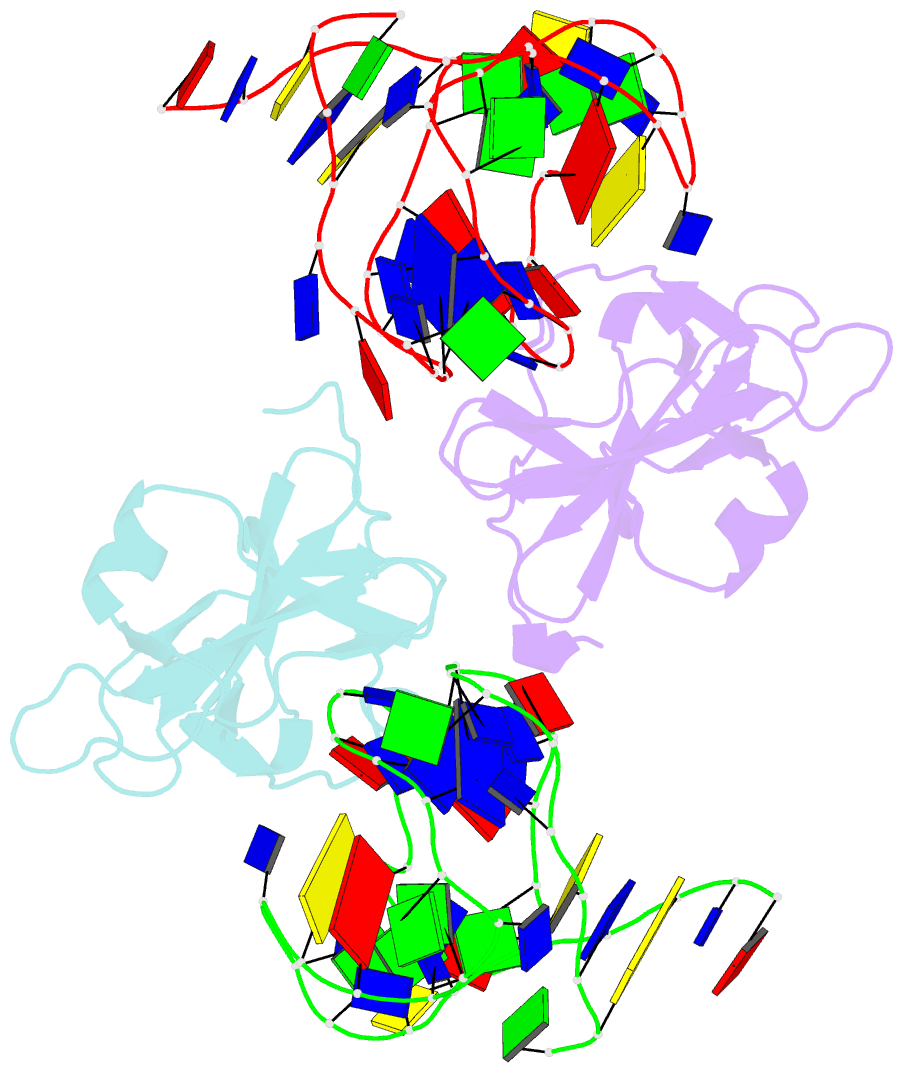
- Structure of a protein-modified aptamer complex
- Reference
- Ren X, Gelinas AD, Linehan M, Iwasaki A, Wang W, Janjic N, Pyle AM (2021): "Evolving A RIG-I Antagonist: A Modified DNA Aptamer Mimics Viral RNA." J.Mol.Biol., 433, 167227. doi: 10.1016/j.jmb.2021.167227.
- Abstract
- Vertebrate organisms express a diversity of protein receptors that recognize and respond to the presence of pathogenic molecules, functioning as an early warning system for infection. As a result of mutation or dysregulated metabolism, these same innate immune receptors can be inappropriately activated, leading to inflammation and disease. One of the most important receptors for detection and response to RNA viruses is called RIG-I, and dysregulation of this protein is linked with a variety of disease states. Despite its central role in inflammatory responses, antagonists for RIG-I are underdeveloped. In this study, we use invitro selection from a pool of modified DNA aptamers to create a high affinity RIG-I antagonist. A high resolution crystal structure of the complex reveals molecular mimicry between the aptamer and the 5'-triphosphate terminus of viral ligands, which bind to the same amino acids within the CTD recognition platform of the RIG-I receptor. Our study suggests a powerful, generalizable strategy for generating immunomodulatory drugs and mechanistic tool compounds.