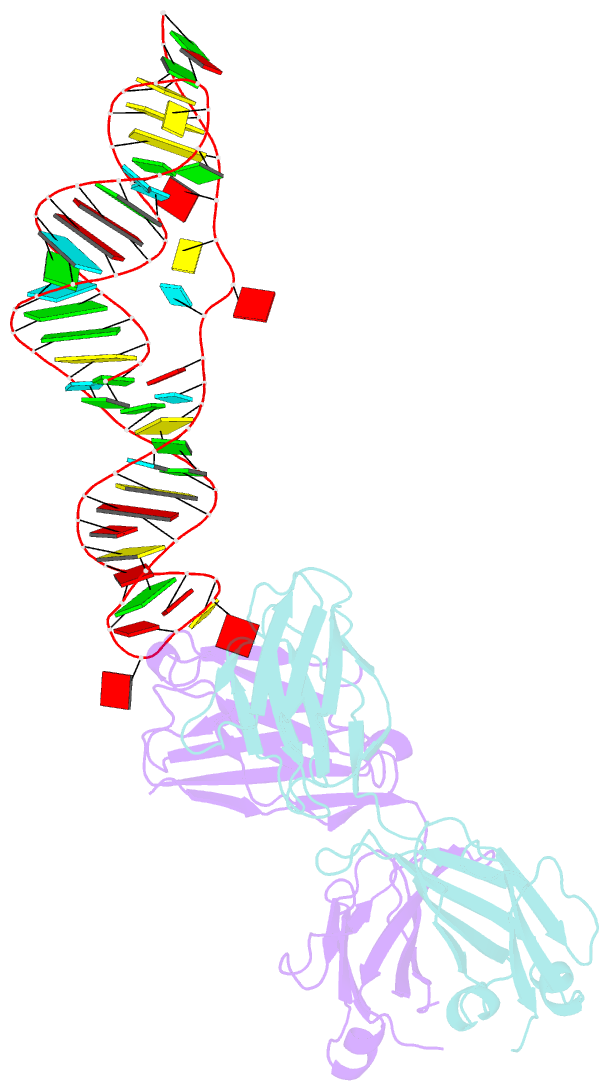
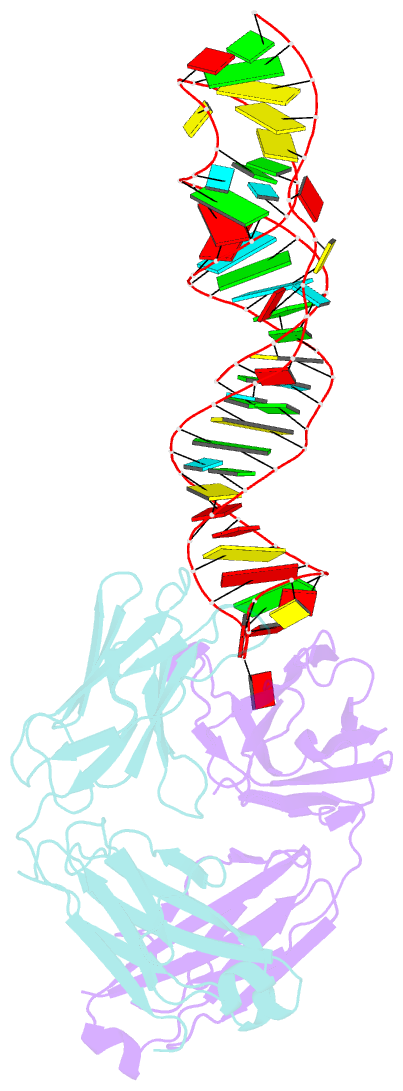
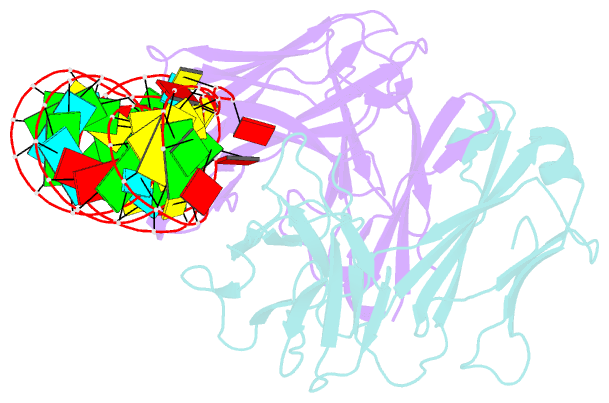
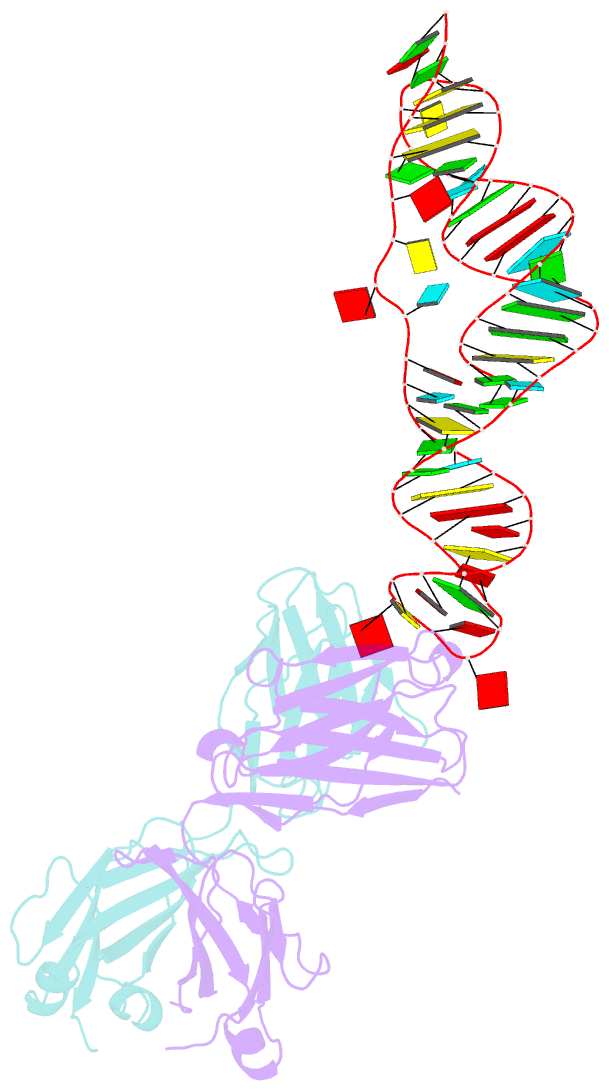
Summary information and primary citation
- PDB-id
- 7mlx; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA-immune system
- Method
- X-ray (2.09 Å)
- Summary
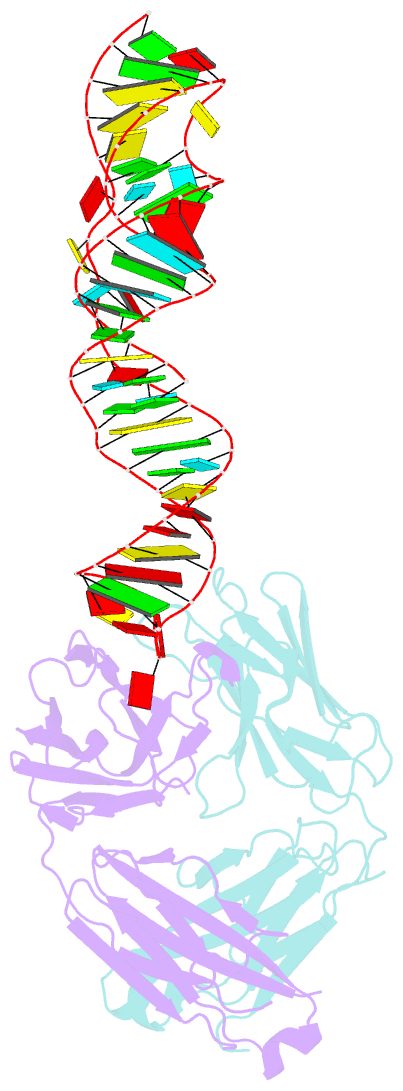
- Sars-cov-2 programmed -1 frameshifting element three stem h-type pseudoknot
- Reference
- Roman C, Lewicka A, Koirala D, Li NS, Piccirilli JA (2021): "The SARS-CoV-2 Programmed -1 Ribosomal Frameshifting Element Crystal Structure Solved to 2.09 angstrom Using Chaperone-Assisted RNA Crystallography." Acs Chem.Biol., 16, 1469-1481. doi: 10.1021/acschembio.1c00324.
- Abstract
- The programmed -1 ribosomal frameshifting element (PFSE) of SARS-CoV-2 is a well conserved structured RNA found in all coronaviruses' genomes. By adopting a pseudoknot structure in the presence of the ribosome, the PFSE promotes a ribosomal frameshifting event near the stop codon of the first open reading frame Orf1a during translation of the polyprotein pp1a. Frameshifting results in continuation of pp1a via a new open reading frame, Orf1b, that produces the longer pp1ab polyprotein. Polyproteins pp1a and pp1ab produce nonstructural proteins NSPs 1-10 and NSPs 1-16, respectively, which contribute vital functions during the viral life cycle and must be present in the proper stoichiometry. Both drugs and sequence alterations that affect the stability of the -1 programmed ribosomal frameshifting element disrupt the stoichiometry of the NSPs produced, which compromise viral replication. For this reason, the -1 programmed frameshifting element is considered a promising drug target. Using chaperone assisted RNA crystallography, we successfully crystallized and solved the three-dimensional structure of the PFSE. We observe a three-stem H-type pseudoknot structure with the three stems stacked in a vertical orientation stabilized by two triple base pairs at the stem 1/stem 2 and stem 1/stem 3 junctions. This structure provides a new conformation of PFSE distinct from the bent conformations inferred from midresolution cryo-EM models and provides a high-resolution framework for mechanistic investigations and structure-based drug design.