Summary information and primary citation
- PDB-id
- 7mrl; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein-RNA
- Method
- X-ray (3.15 Å)
- Summary
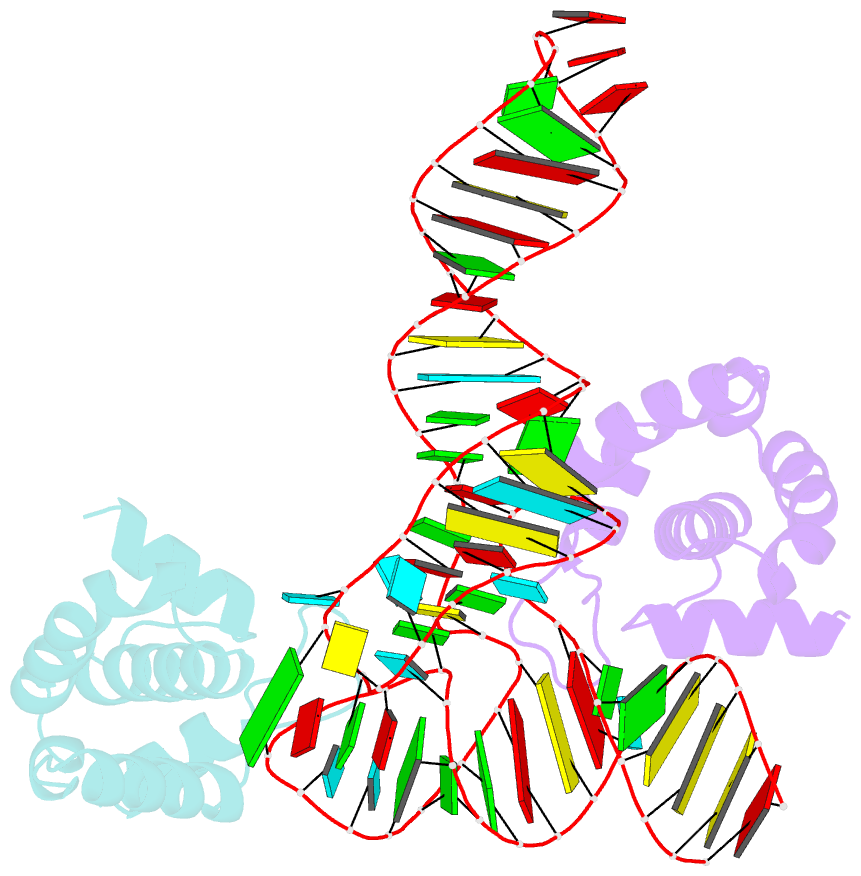
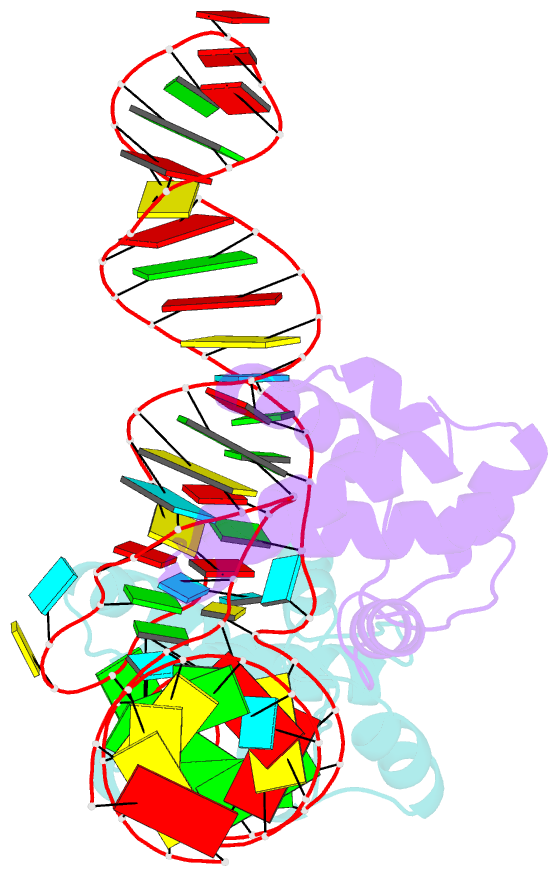
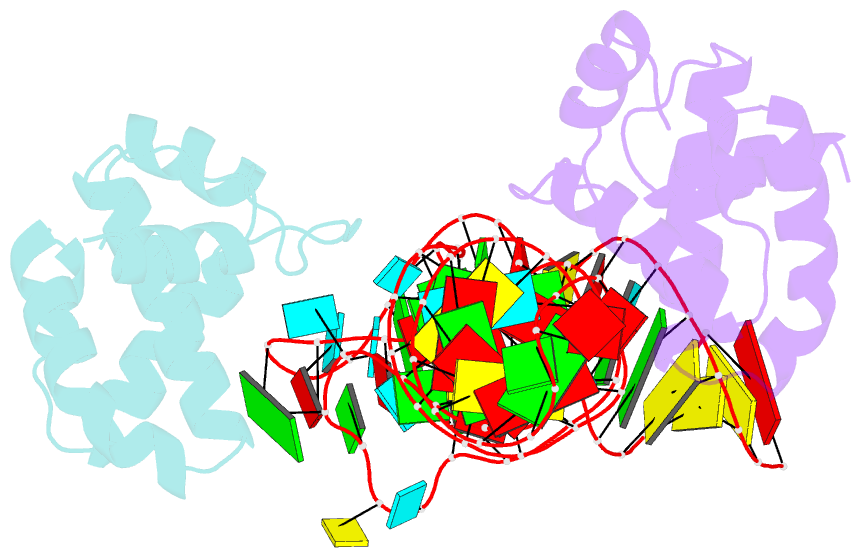
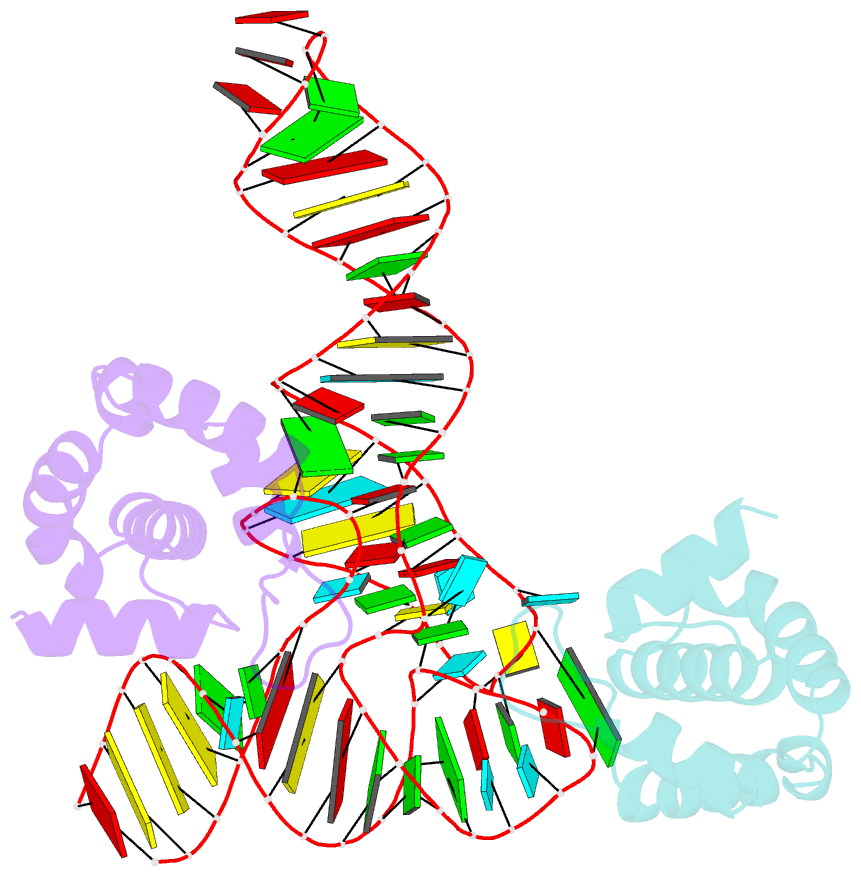
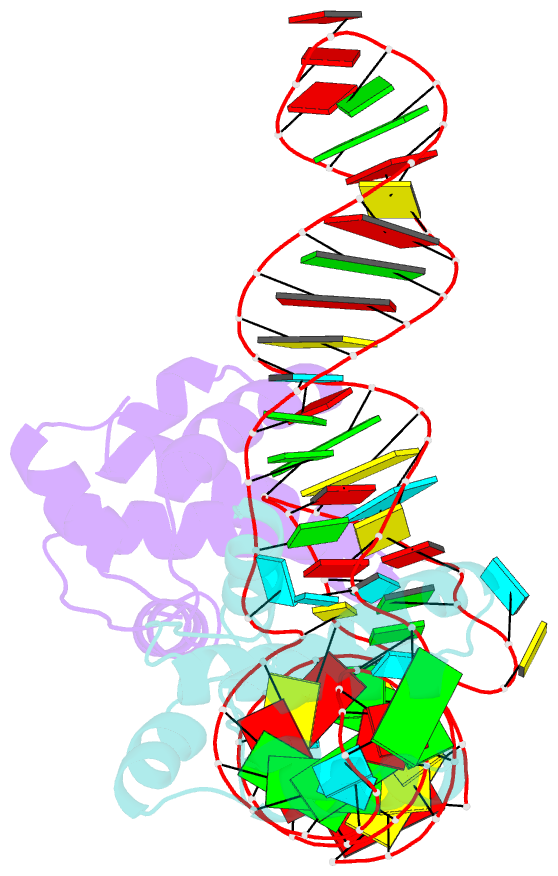
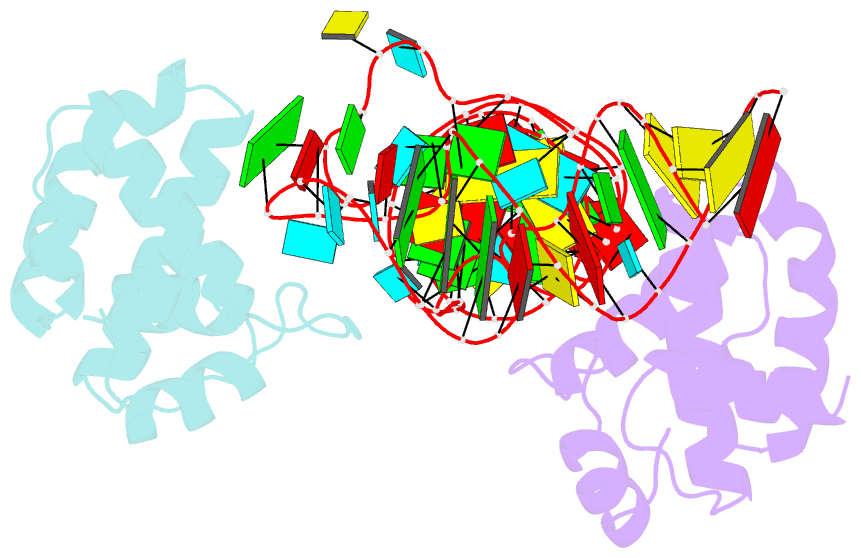
- Structure of hiv-1 matrix domain bound to human trnalys3
- Reference
- Bou-Nader C, Muecksch F, Brown JB, Gordon JM, York A, Peng C, Ghirlando R, Summers MF, Bieniasz PD, Zhang J (2021): "HIV-1 matrix-tRNA complex structure reveals basis for host control of Gag localization." Cell Host Microbe, 29, 1421. doi: 10.1016/j.chom.2021.07.006.
- Abstract
- The HIV-1 virion structural polyprotein, Gag, is directed to particle assembly sites at the plasma membrane by its N-terminal matrix (MA) domain. MA also binds to host tRNAs. To understand the molecular basis of MA-tRNA interaction and its potential function, we present a co-crystal structure of HIV-1 MA-tRNALys3 complex. The structure reveals a specialized group of MA basic and aromatic residues preconfigured to recognize the distinctive structure of the tRNA elbow. Mutational, cross-linking, fluorescence, and NMR analyses show that the crystallographically defined interface drives MA-tRNA binding in solution and living cells. The structure indicates that MA is unlikely to bind tRNA and membrane simultaneously. Accordingly, single-amino-acid substitutions that abolish MA-tRNA binding caused striking redistribution of Gag to the plasma membrane and reduced HIV-1 replication. Thus, HIV-1 exploits host tRNAs to occlude a membrane localization signal and control the subcellular distribution of its major structural protein.