Summary information and primary citation
- PDB-id
- 7mtl; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.446 Å)
- Summary
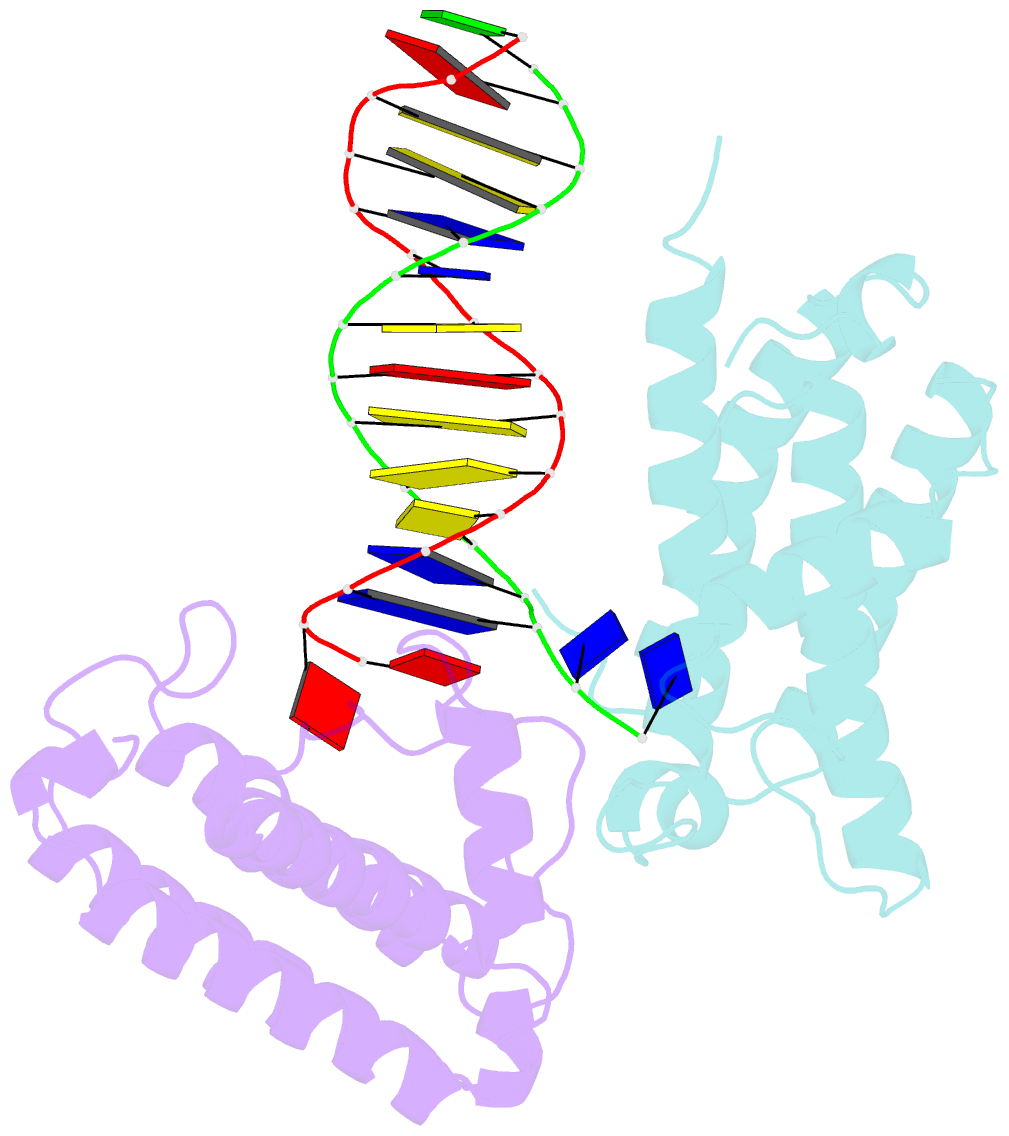
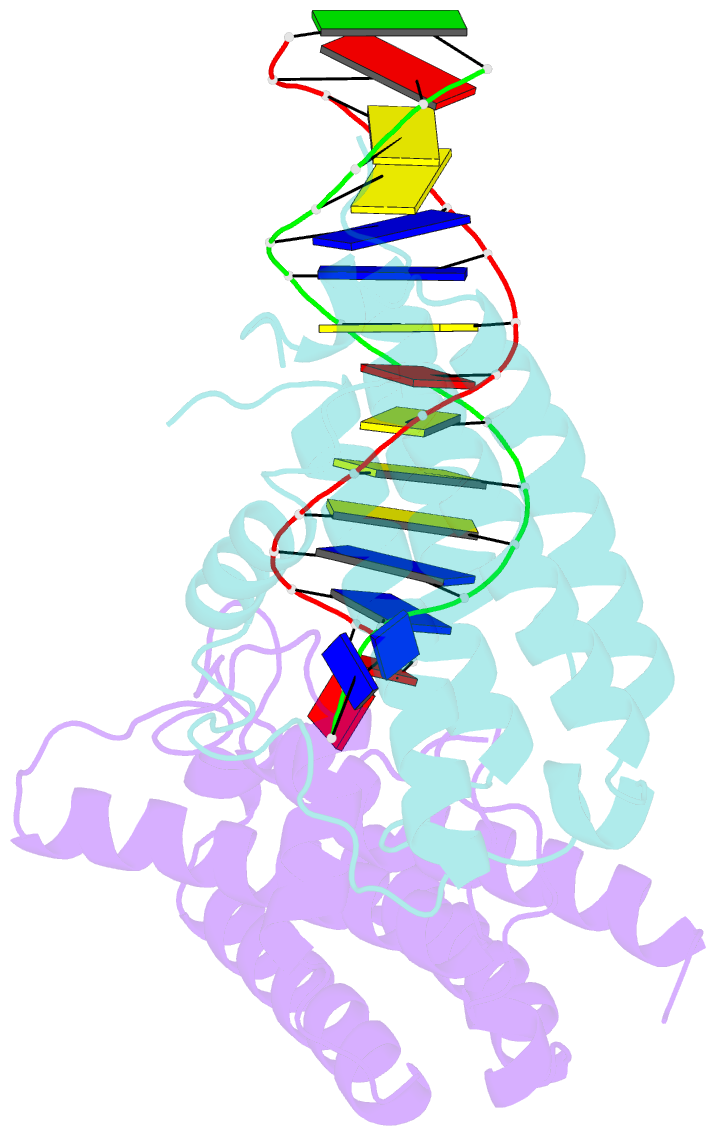
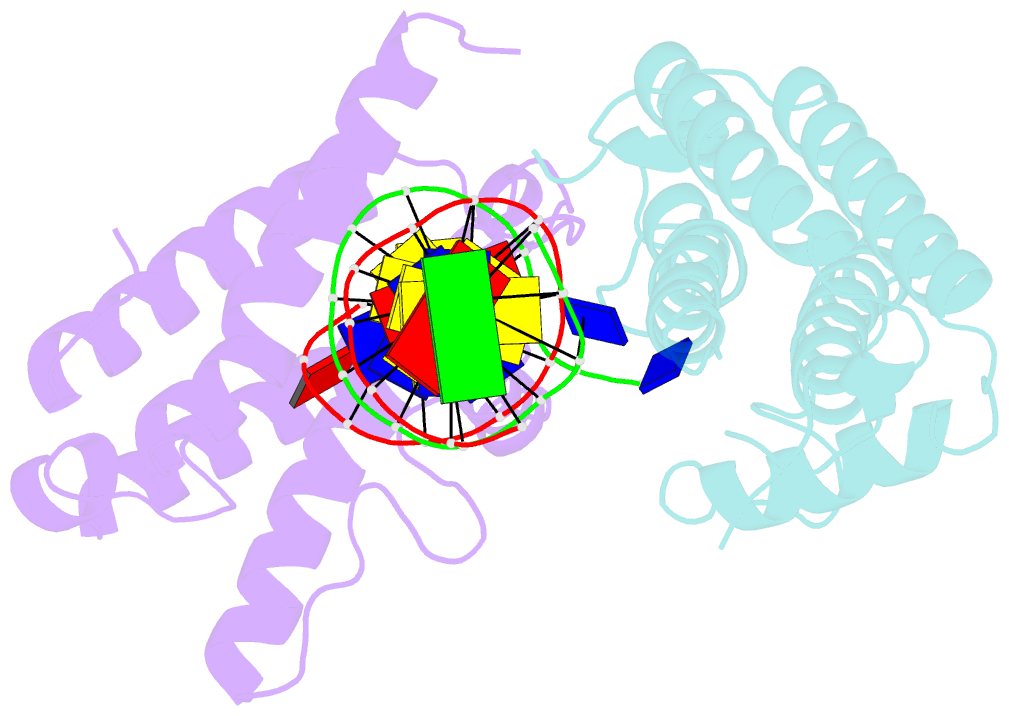
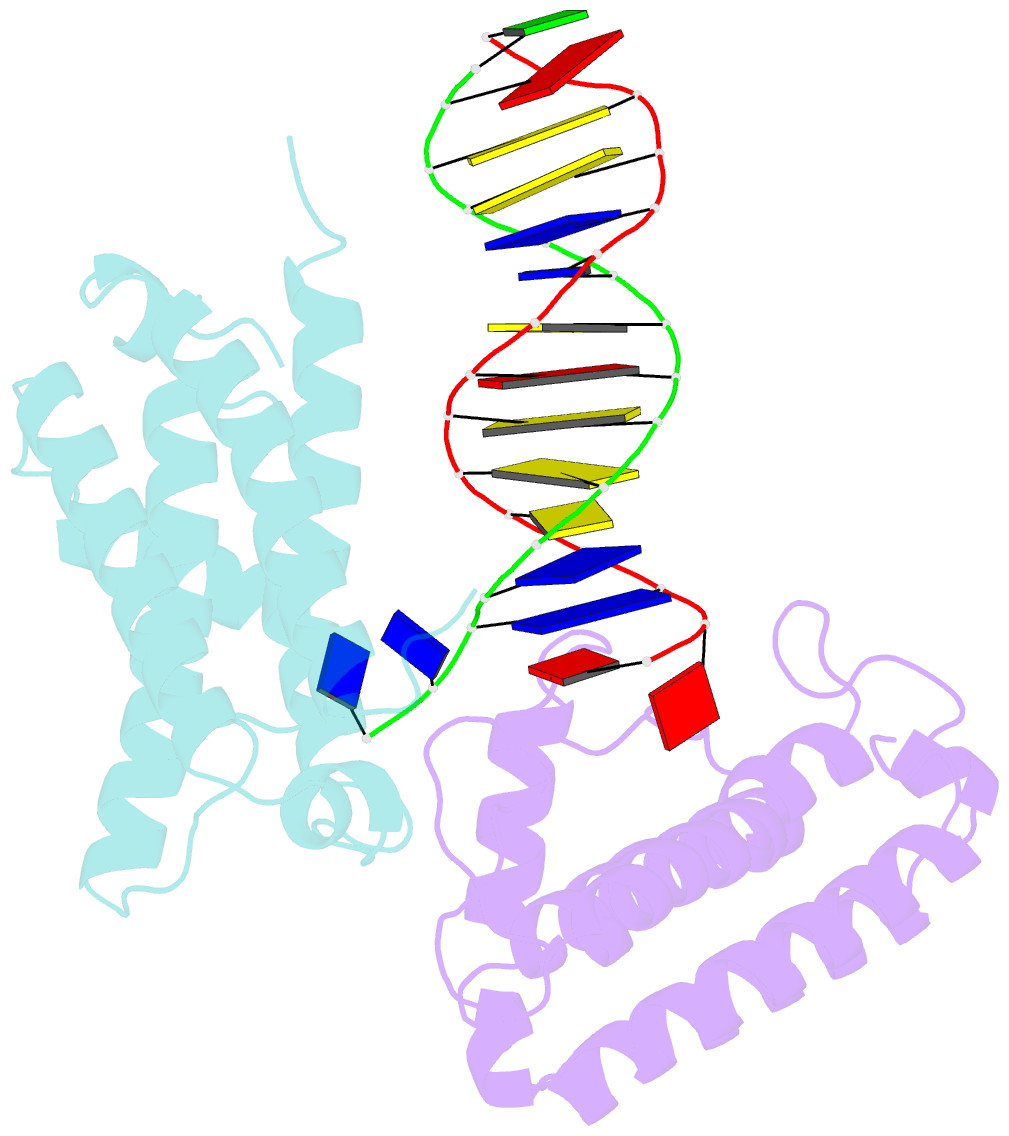
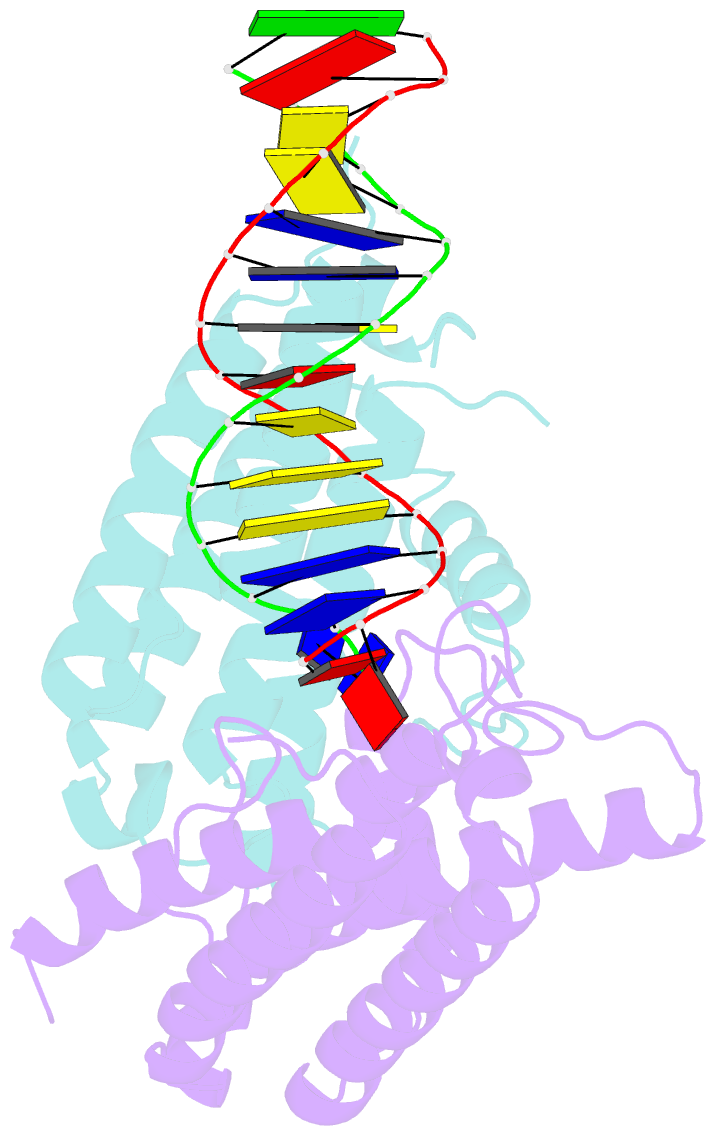
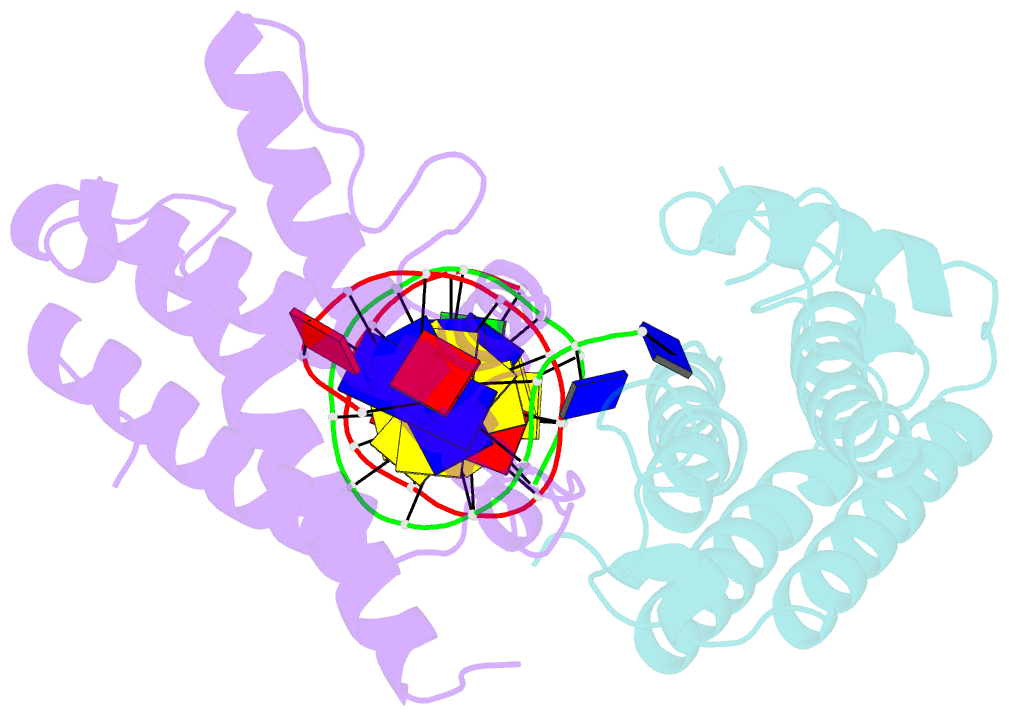
- Crystal structure of colibactin self-resistance protein clbs in complex with a dsDNA
- Reference
- Tripathi P, Bruner SD (2021): "Structural Basis for the Interactions of the Colibactin Resistance Gene Product ClbS with DNA." Biochemistry, 60, 1619-1625. doi: 10.1021/acs.biochem.1c00201.
- Abstract
- The natural product colibactin, along with its associated biosynthetic gene cluster, is an example system for the role microbially derived small molecules play in the human microbiome. This is particularly relevant in the human gut, where host microbiota is involved in various disorders, including colorectal cancer pathogenesis. Bacteria harboring the colibactin gene cluster induce alkylation of nucleobases in host DNA, forming interstrand cross-links both in vivo and in vitro. These lesions can lead to deleterious double-strand breaks and have been identified as the primary mechanism of colibactin-induced cytotoxicity. The gene product ClbS is one of several mechanisms utilized by the producing bacteria to maintain genome integrity. ClbS catalyzes hydrolytic inactivation of colibactin and has been shown to bind DNA, incurring self-resistance. Presented is the molecular basis for ClbS bound to a DNA oligonucleotide. The structure shows the interaction of the protein with the ends of a DNA duplex with terminal nucleotides flipped to the enzyme active site. The structure suggests an additional function for ClbS, the binding to damaged DNA followed by repair. Additionally, our study provides general insight into the function of the widely distributed and largely uncharacterized DUF1706 protein family.