Summary information and primary citation
- PDB-id
- 7n2m; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA-RNA
- Method
- X-ray (2.9 Å)
- Summary
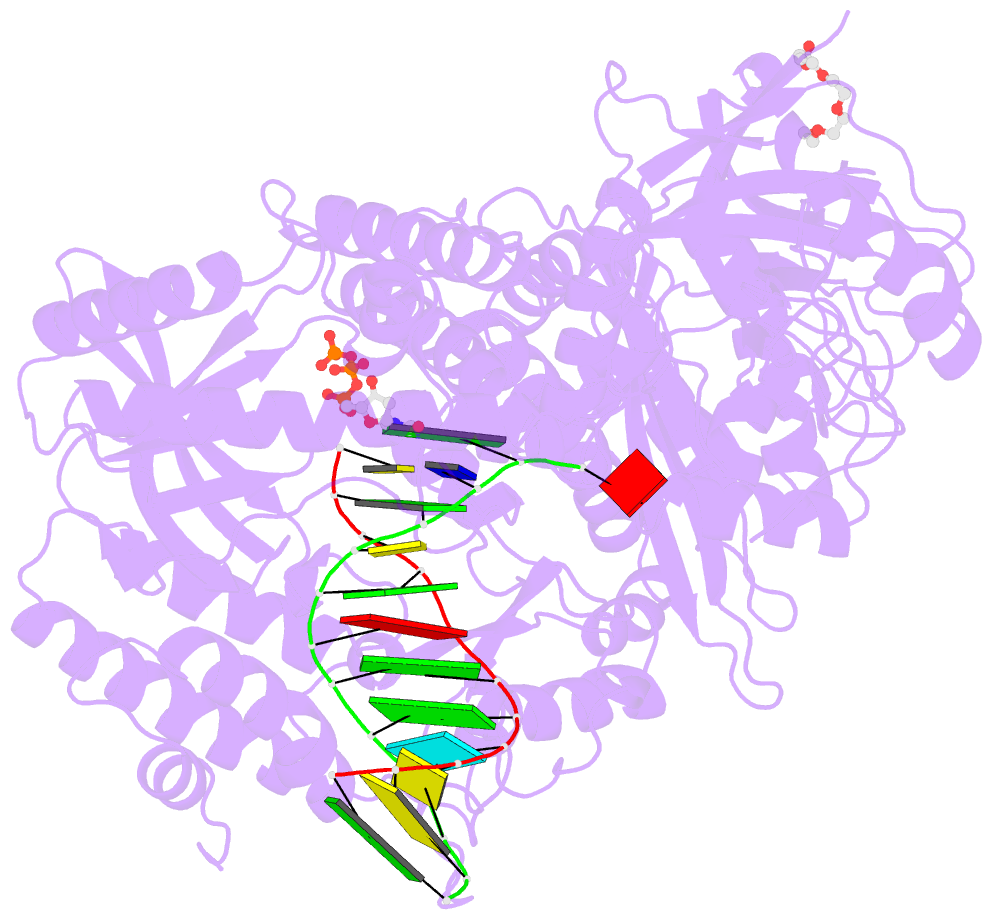
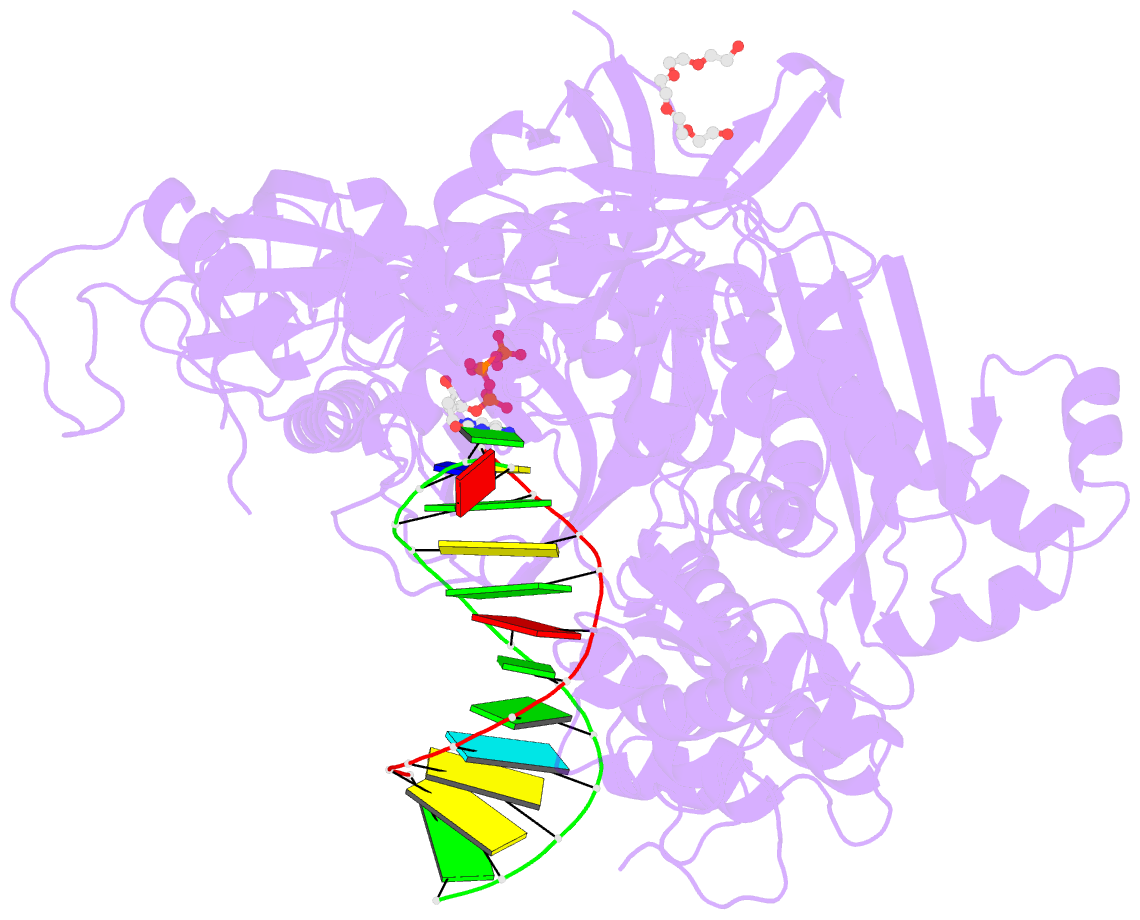
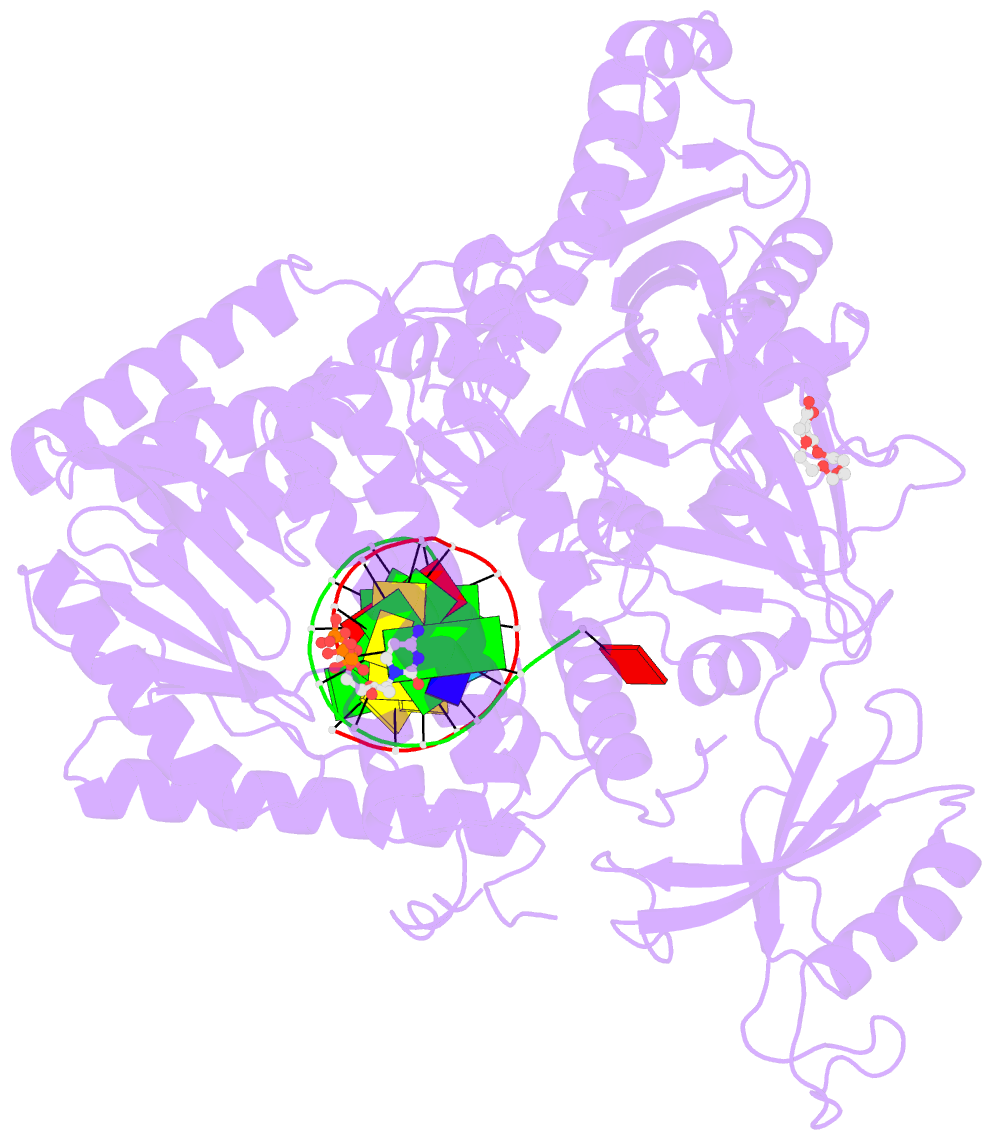
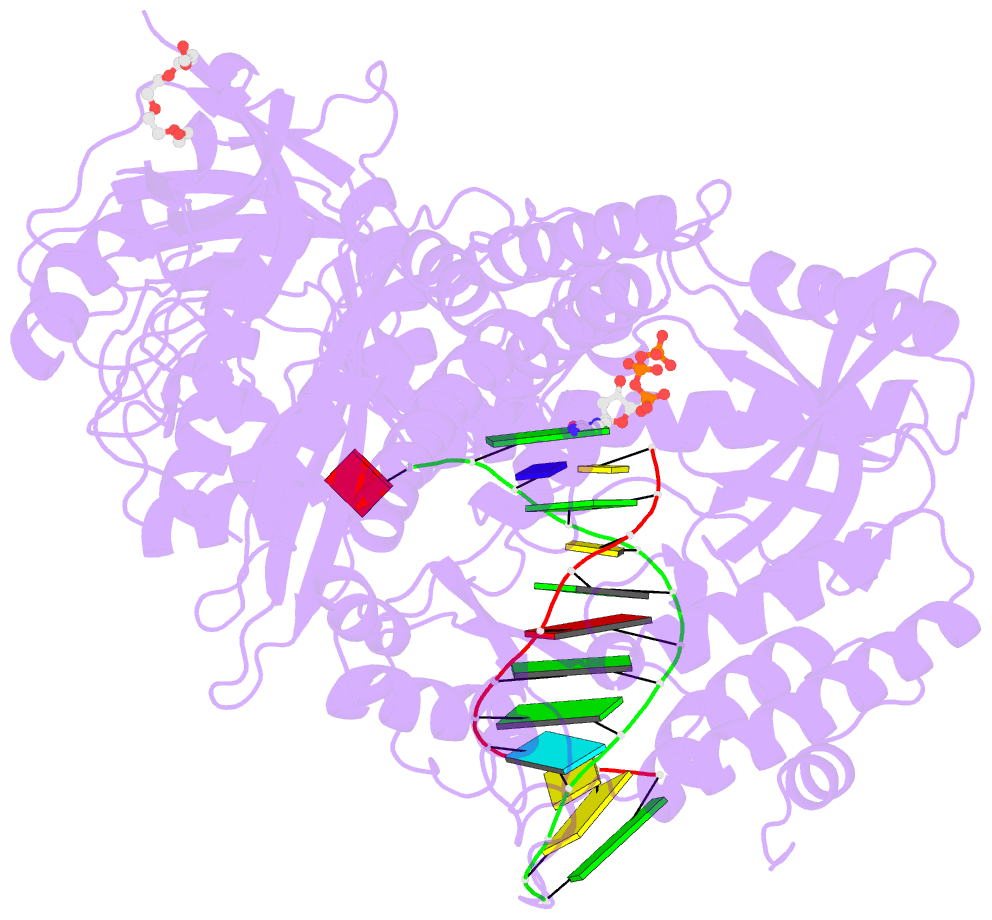
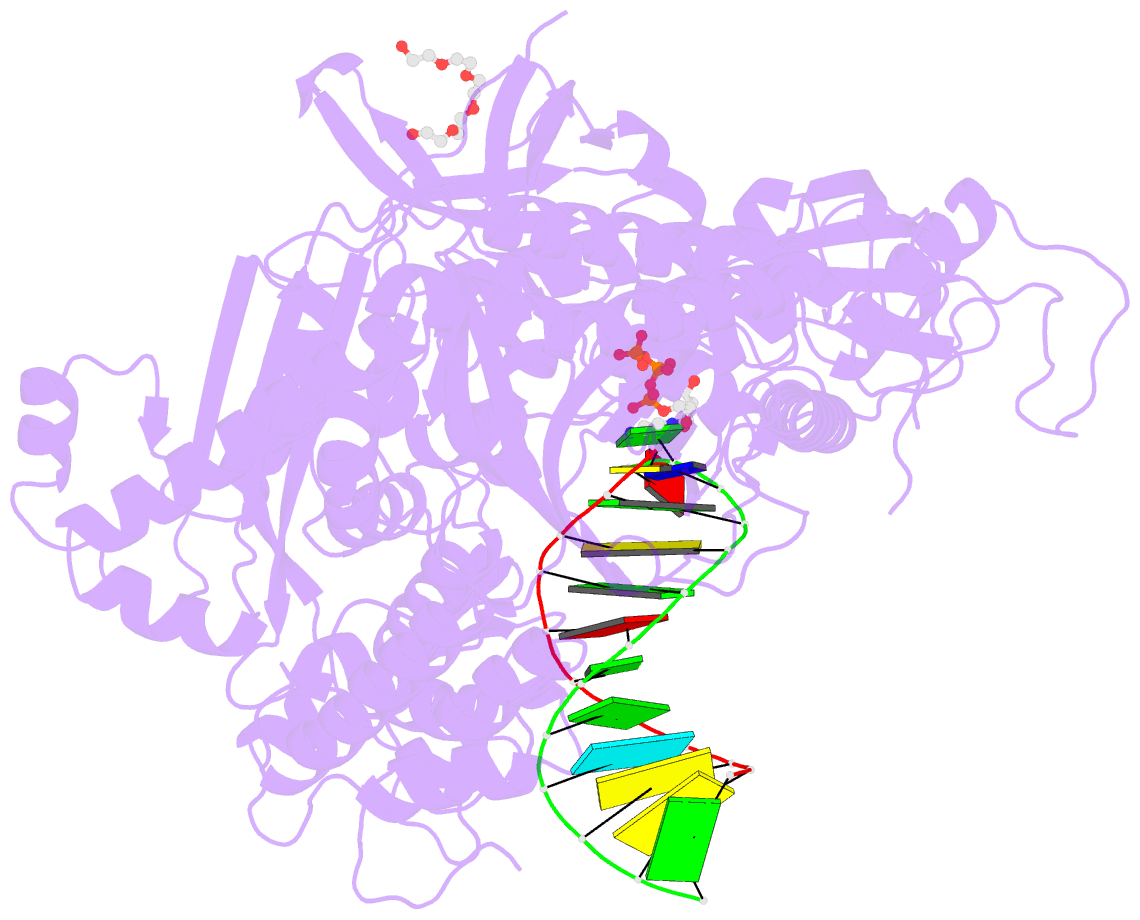
- Crystal structure of DNA polymerase alpha catalytic core in complex with dctp and template-primer having t-c mismatch at the post-insertion site
- Reference
- Baranovskiy AG, Babayeva ND, Lisova AE, Morstadt LM, Tahirov TH (2022): "Structural and functional insight into mismatch extension by human DNA polymerase alpha." Proc.Natl.Acad.Sci.USA, 119, e2111744119. doi: 10.1073/pnas.2111744119.
- Abstract
- Human DNA polymerase α (Polα) does not possess proofreading ability and plays an important role in genome replication and mutagenesis. Polα extends the RNA primers generated by primase and provides a springboard for loading other replication factors. Here we provide the structural and functional analysis of the human Polα interaction with a mismatched template:primer. The structure of the human Polα catalytic domain in the complex with an incoming deoxycytidine triphosphate (dCTP) and the template:primer containing a T-C mismatch at the growing primer terminus was solved at a 2.9 Å resolution. It revealed the absence of significant distortions in the active site and in the conformation of the substrates, except the primer 3′-end. The T-C mismatch acquired a planar geometry where both nucleotides moved toward each other by 0.4 Å and 0.7 Å, respectively, and made one hydrogen bond. The binding studies conducted at a physiological salt concentration revealed that Polα has a low affinity to DNA and is not able to discriminate against a mispaired template:primer in the absence of deoxynucleotide triphosphate (dNTP). Strikingly, in the presence of cognate dNTP, Polα showed a more than 10-fold higher selectivity for a correct duplex versus a mismatched one. According to pre-steady-state kinetic studies, human Polα extends the T-C mismatch with a 249-fold lower efficiency due to reduction of the polymerization rate constant by 38-fold and reduced affinity to the incoming nucleotide by 6.6-fold. Thus, a mismatch at the postinsertion site affects all factors important for primer extension: affinity to both substrates and the rate of DNA polymerization.