Summary information and primary citation
- PDB-id
- 7n3o; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- cryo-EM (3.8 Å)
- Summary
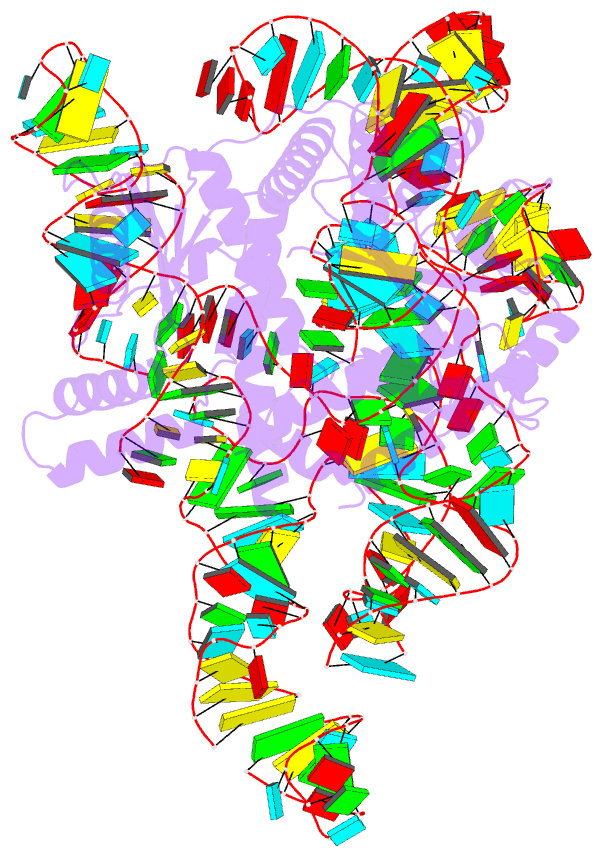
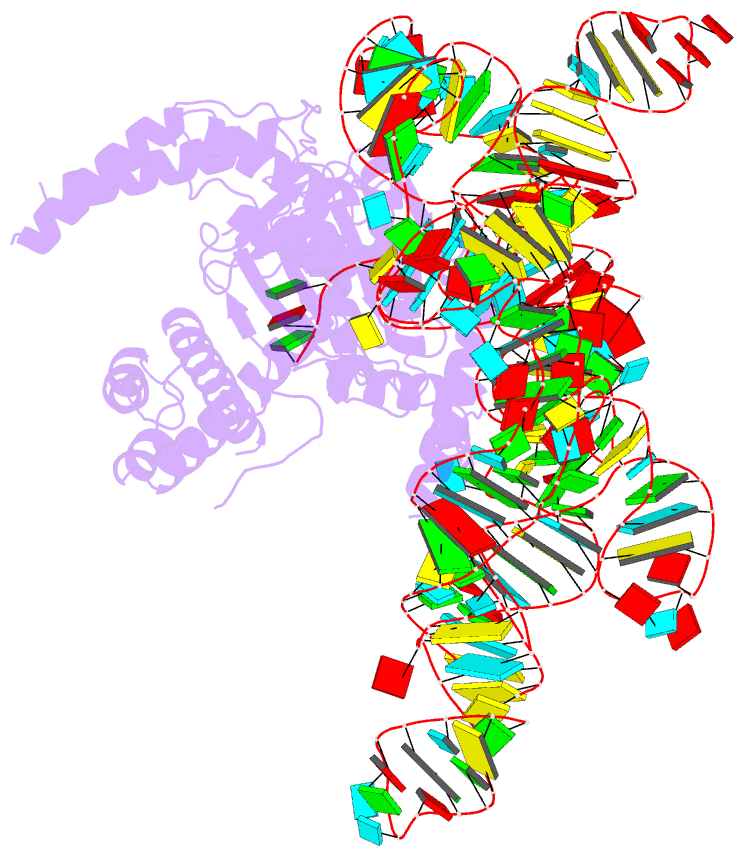
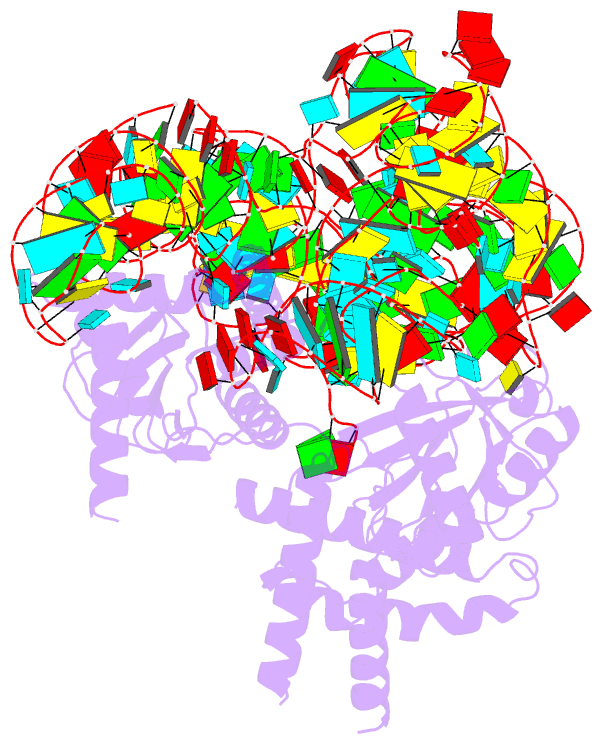
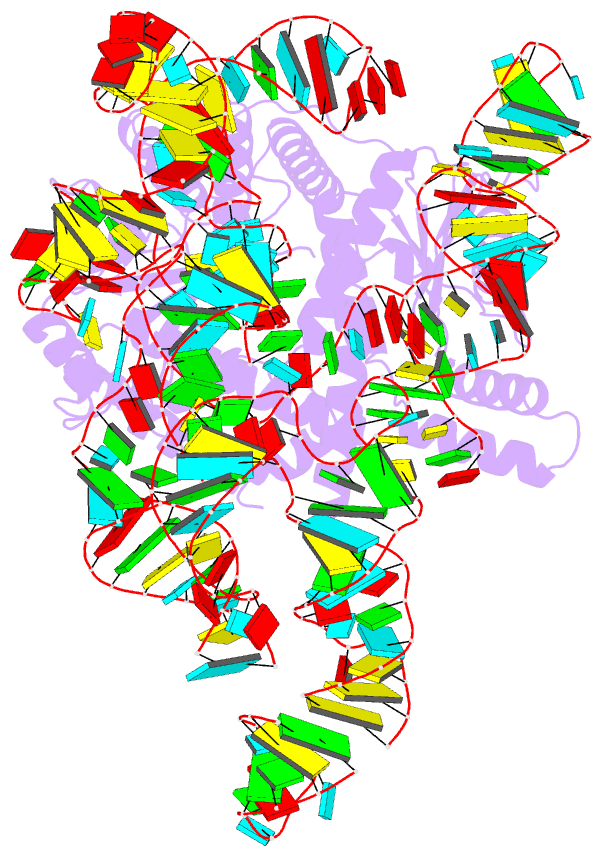
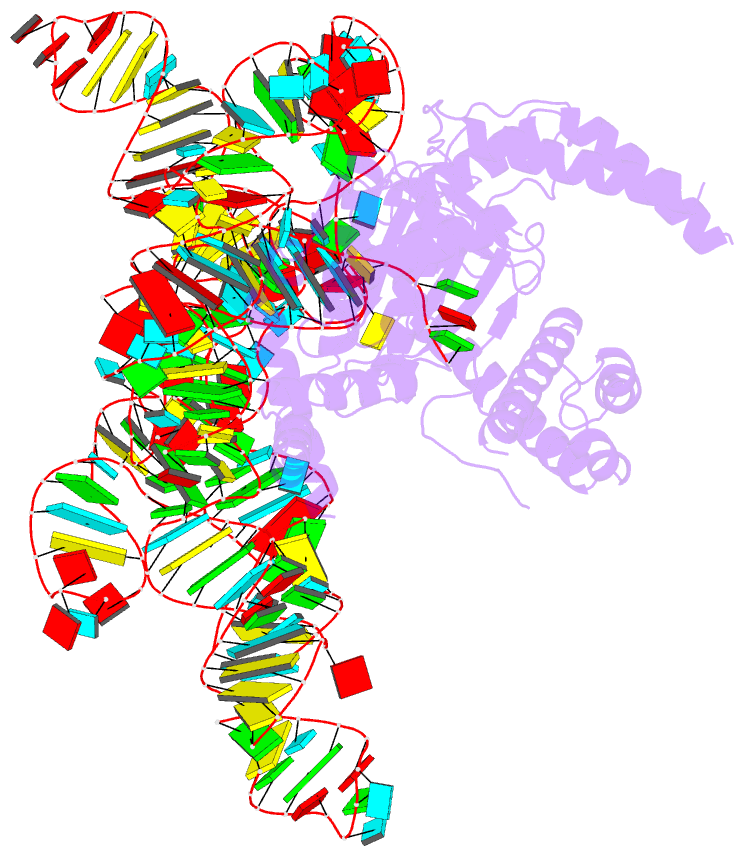
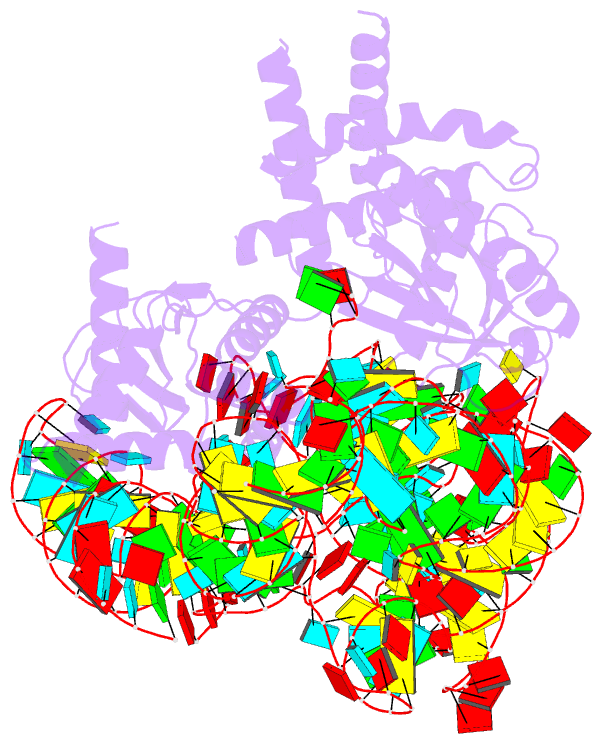
- cryo-EM structure of the cas12k-sgrna complex
- Reference
- Xiao R, Wang S, Han R, Li Z, Gabel C, Mukherjee IA, Chang L (2021): "Structural basis of target DNA recognition by CRISPR-Cas12k for RNA-guided DNA transposition." Mol.Cell, 81, 4457-4466.e5. doi: 10.1016/j.molcel.2021.07.043.
- Abstract
- The type V-K CRISPR-Cas system, featured by Cas12k effector with a naturally inactivated RuvC domain and associated with Tn7-like transposon for RNA-guided DNA transposition, is a promising tool for precise DNA insertion. To reveal the mechanism underlying target DNA recognition, we determined a cryoelectron microscopy (cryo-EM) structure of Cas12k from cyanobacteria Scytonema hofmanni in complex with a single guide RNA (sgRNA) and a double-stranded target DNA. Coupled with mutagenesis and in vitro DNA transposition assay, our results revealed mechanisms for the recognition of the GGTT protospacer adjacent motif (PAM) sequence and the structural elements of Cas12k critical for RNA-guided DNA transposition. These structural and mechanistic insights should aid in the development of type V-K CRISPR-transposon systems as tools for genome editing.