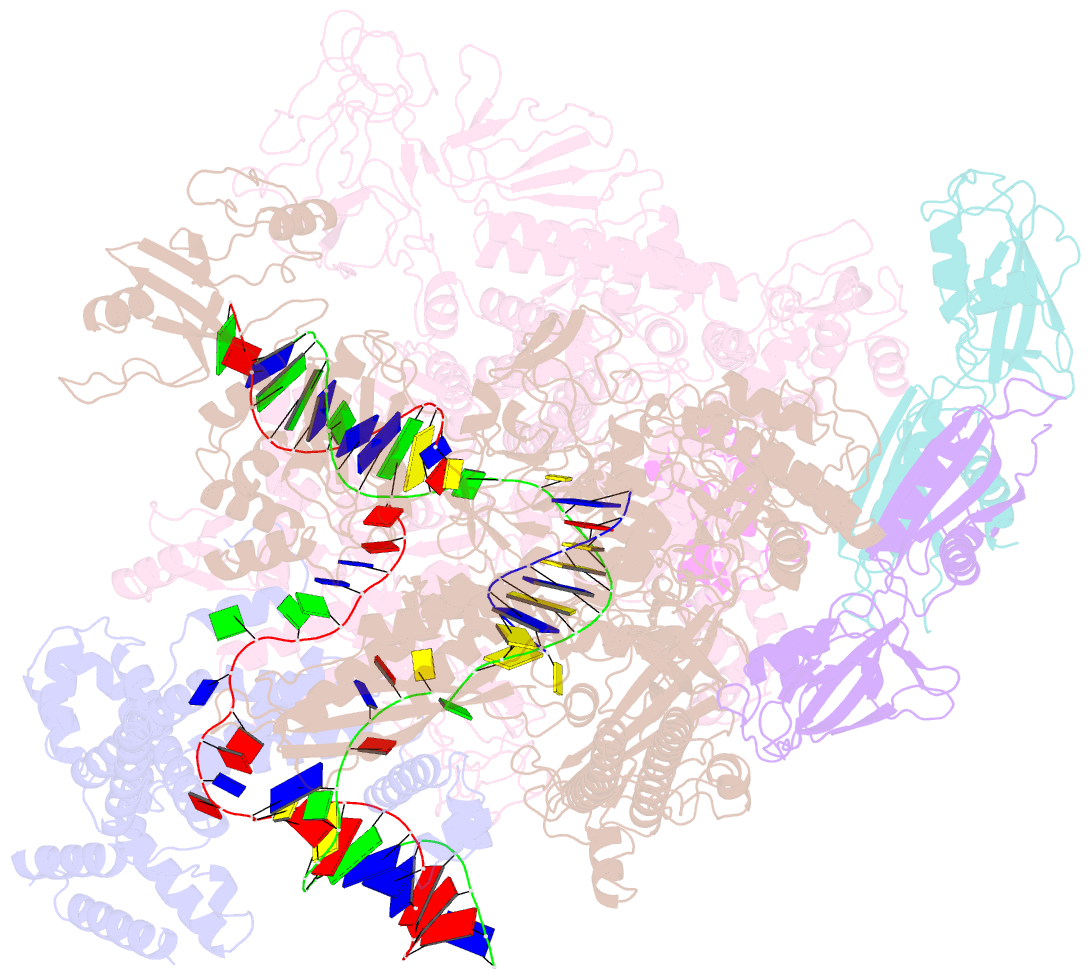
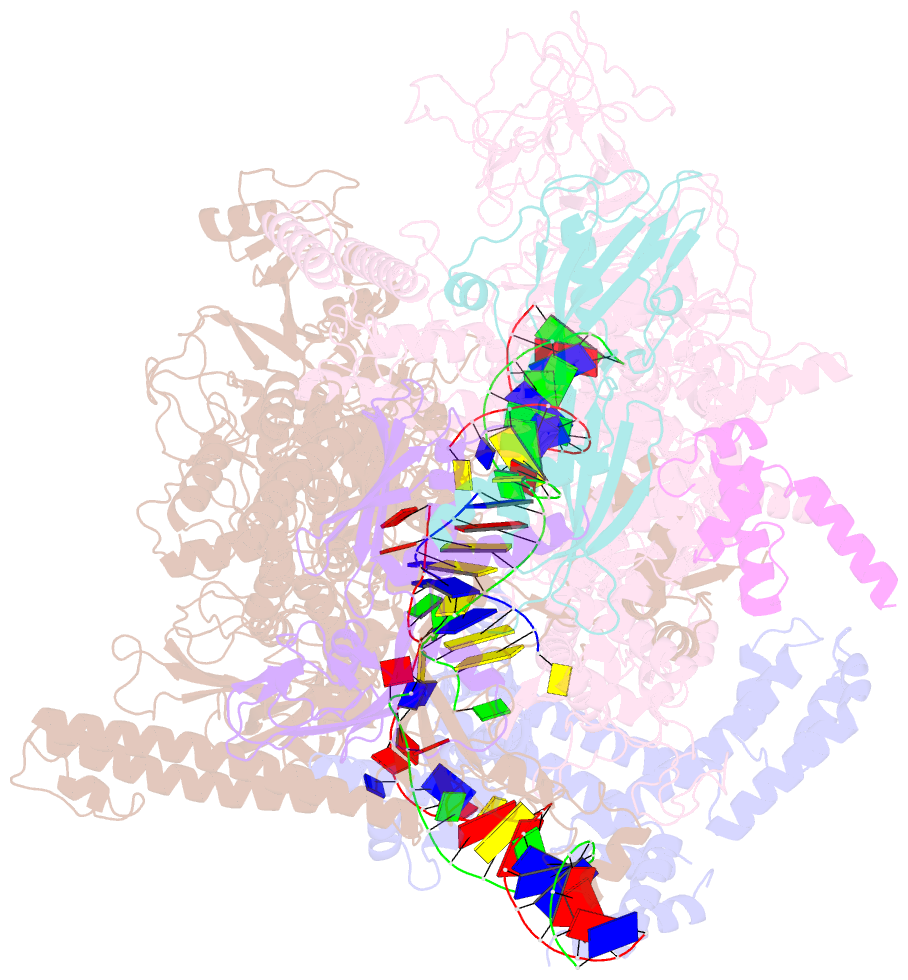
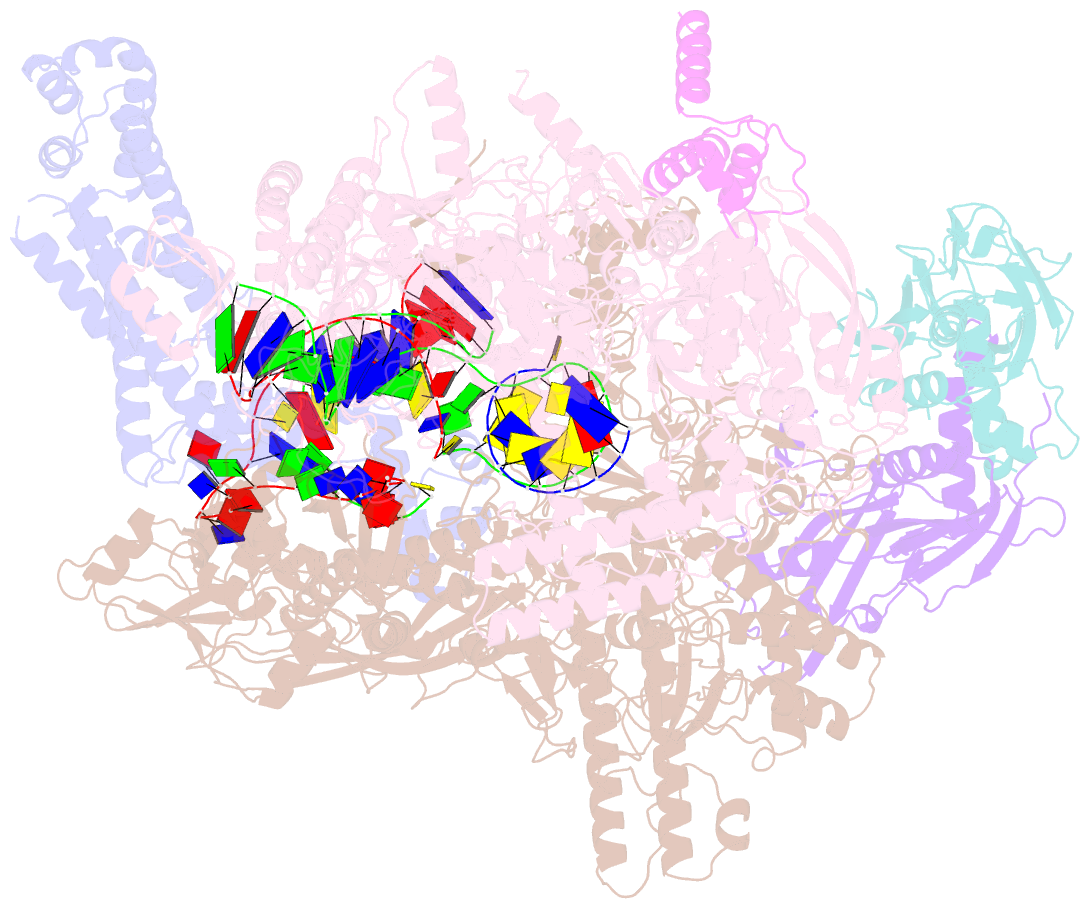
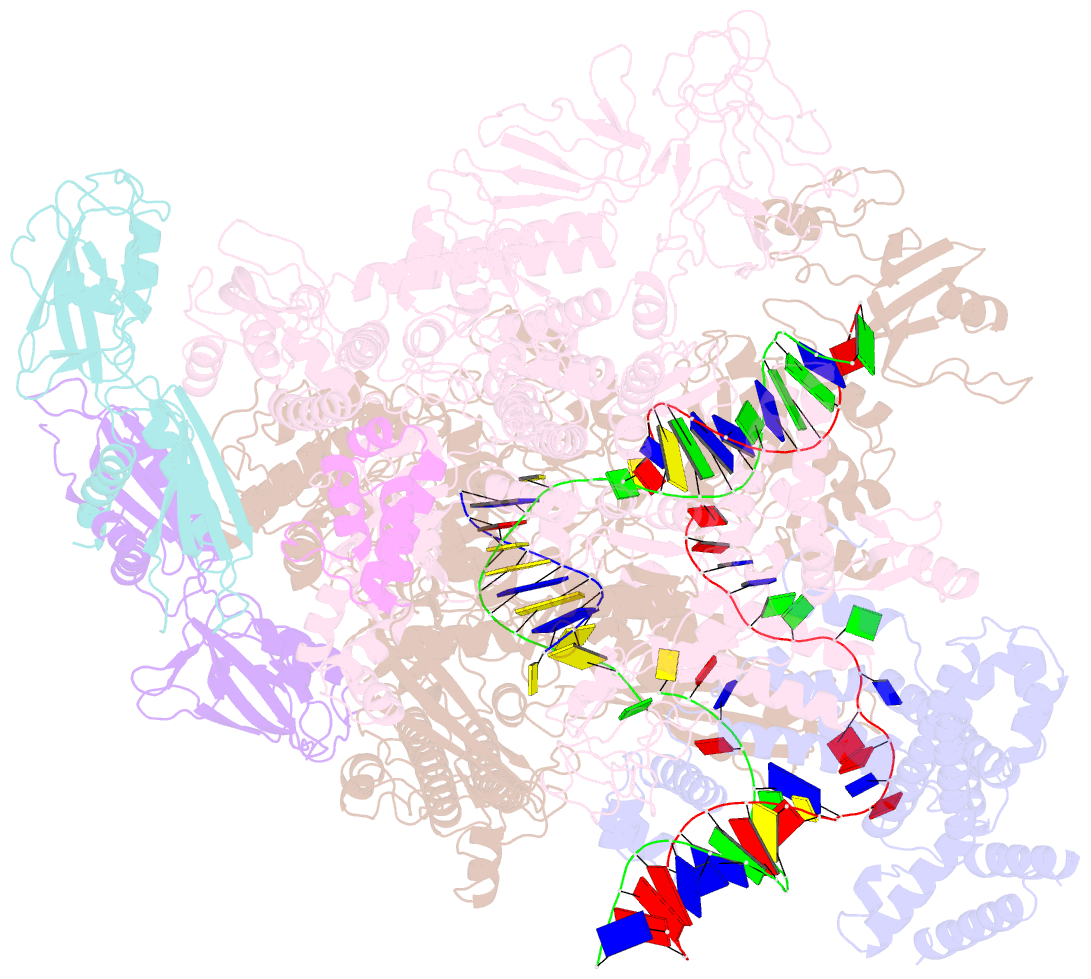
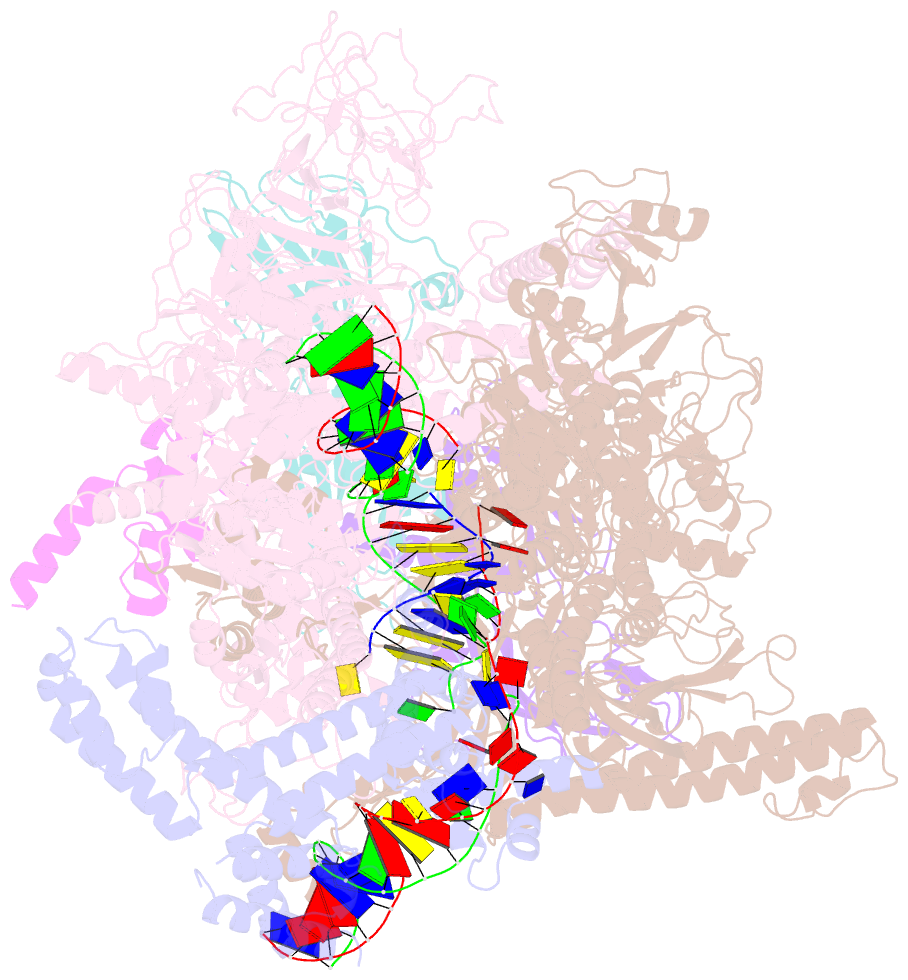
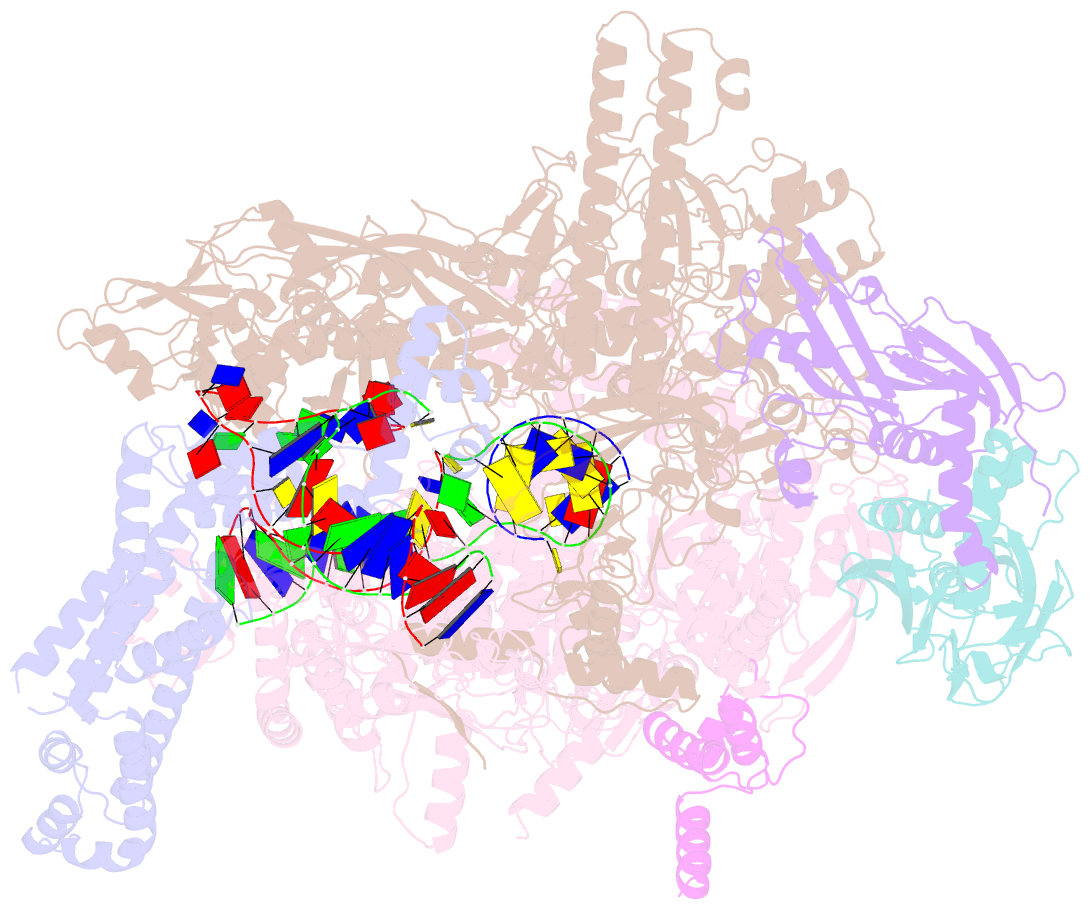
Summary information and primary citation
- PDB-id
- 7n4e; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA-RNA
- Method
- cryo-EM (3.8 Å)
- Summary
- Escherichia coli sigma 70-dependent paused transcription elongation complex
- Reference
- Pukhrambam C, Molodtsov V, Kooshkbaghi M, Tareen A, Vu H, Skalenko KS, Su M, Yin Z, Winkelman JT, Kinney JB, Ebright RH, Nickels BE (2022): "Structural and mechanistic basis of sigma-dependent transcriptional pausing." Proc.Natl.Acad.Sci.USA, 119, e2201301119. doi: 10.1073/pnas.2201301119.
- Abstract
- SignificanceThe paradigmatic example of factor-dependent pausing in transcription elongation is σ-dependent pausing, in which sequence-specific σ-DNA interaction with a - 10 element-like sequence in a transcribed region results in pausing of a σ-containing transcription elongation complex. It has been proposed that σ-dependent pausing involves DNA scrunching, and that sequences downstream of the -10 element-like sequence modulate DNA scrunching. Here, using site-specific protein-DNA photocrosslinking, high-throughput sequencing, and cryoelectron microscopy structure determination, we show directly that σ-dependent pausing involves DNA scrunching, we define a consensus sequence for formation of a stable scrunched paused complex that is identical to the consensus sequence for pausing in initial transcription, and we identify positions of DNA scrunching on DNA nontemplate and template strands. Our results illuminate the structural and mechanistic basis of σ-dependent transcriptional pausing.