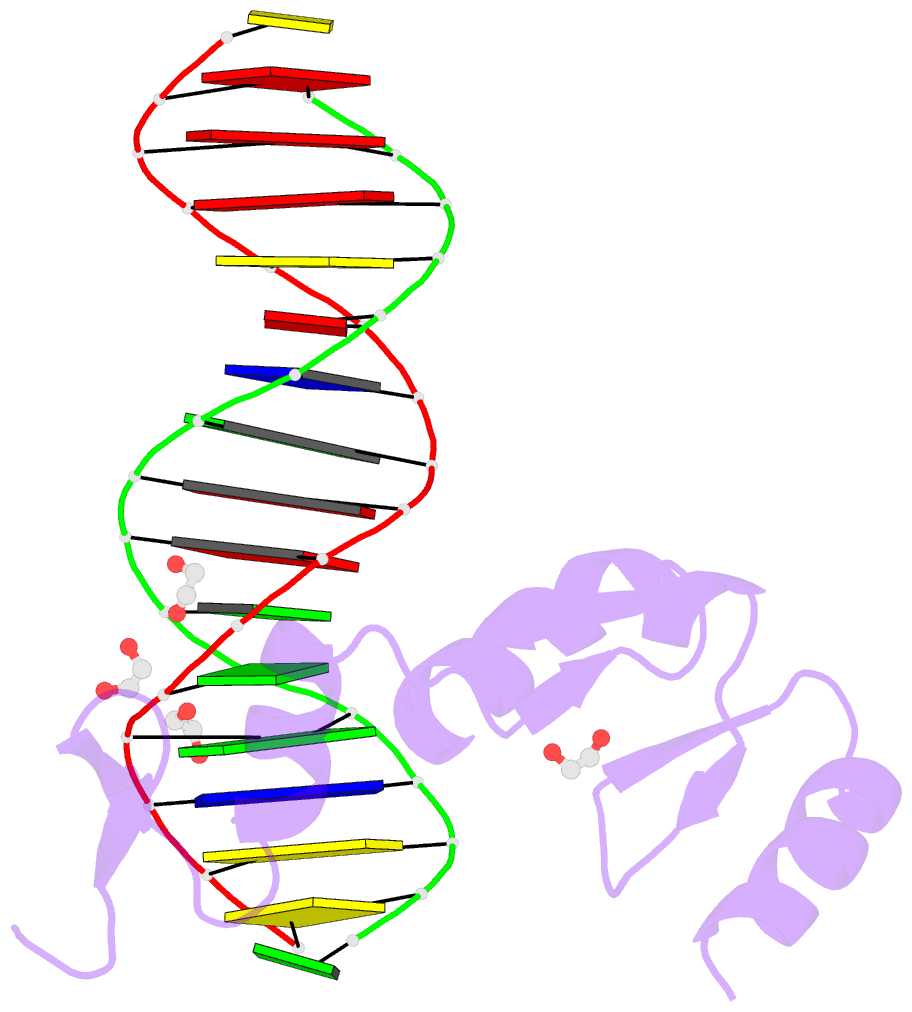
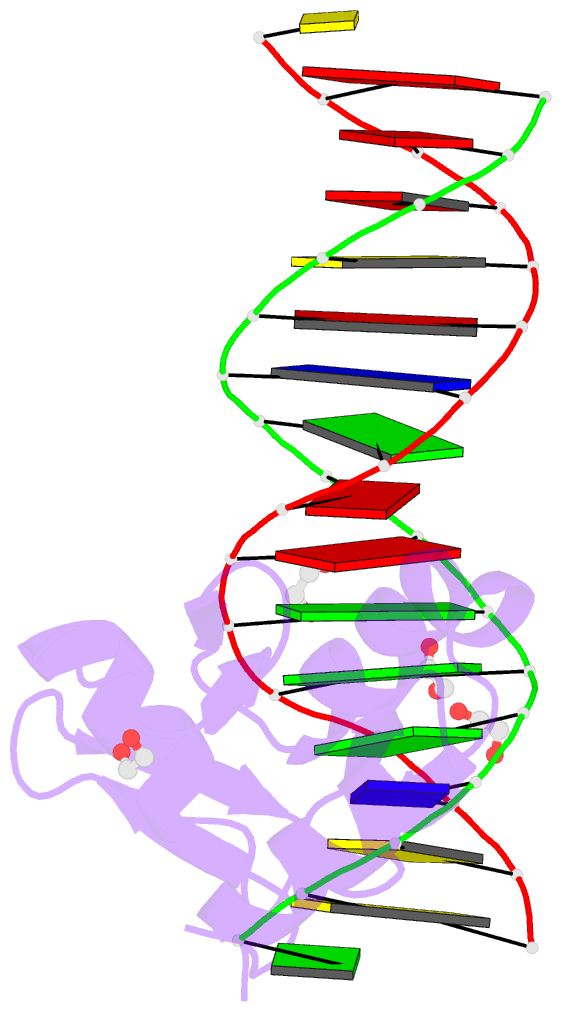
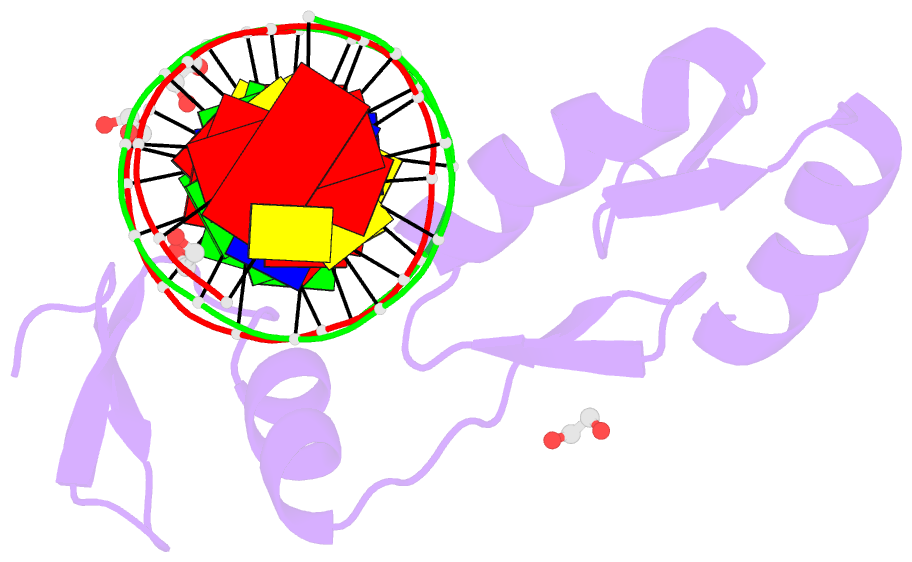
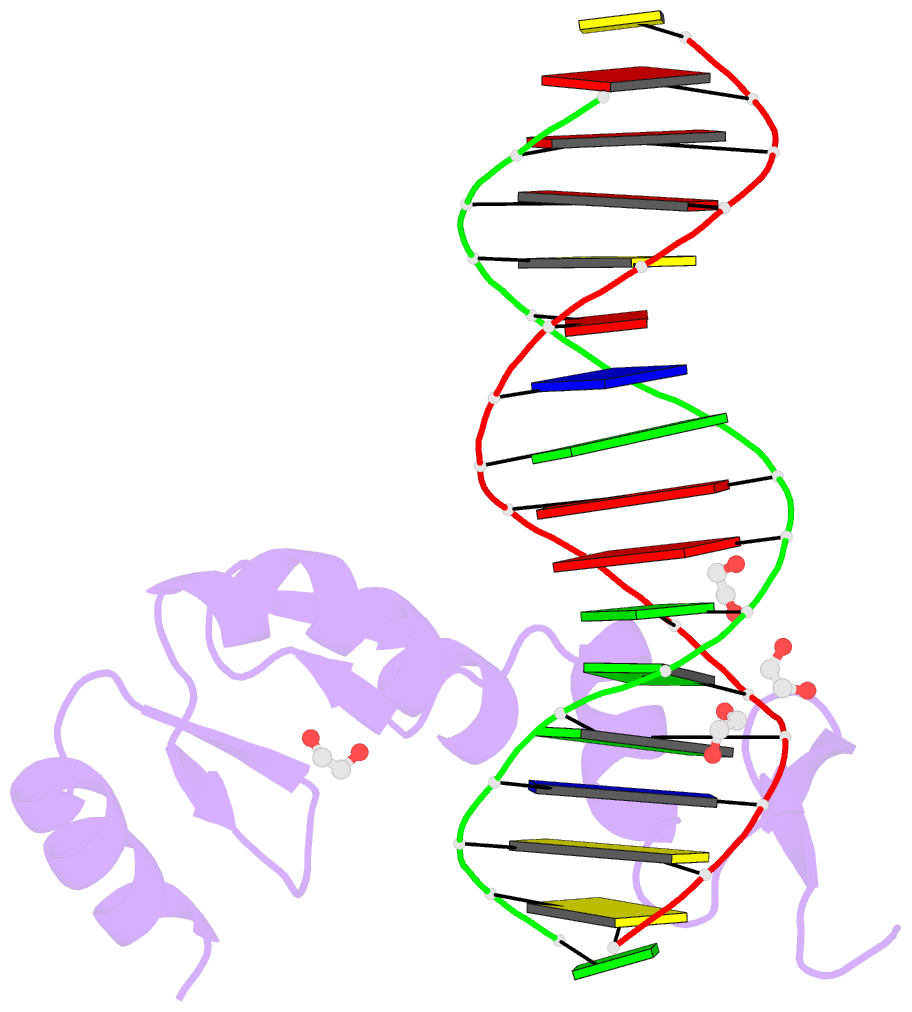
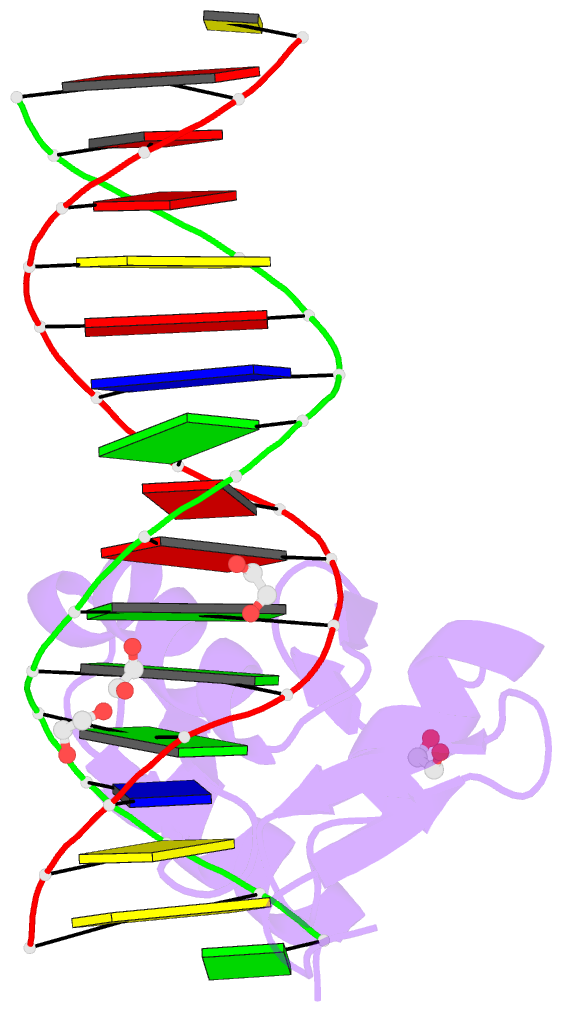
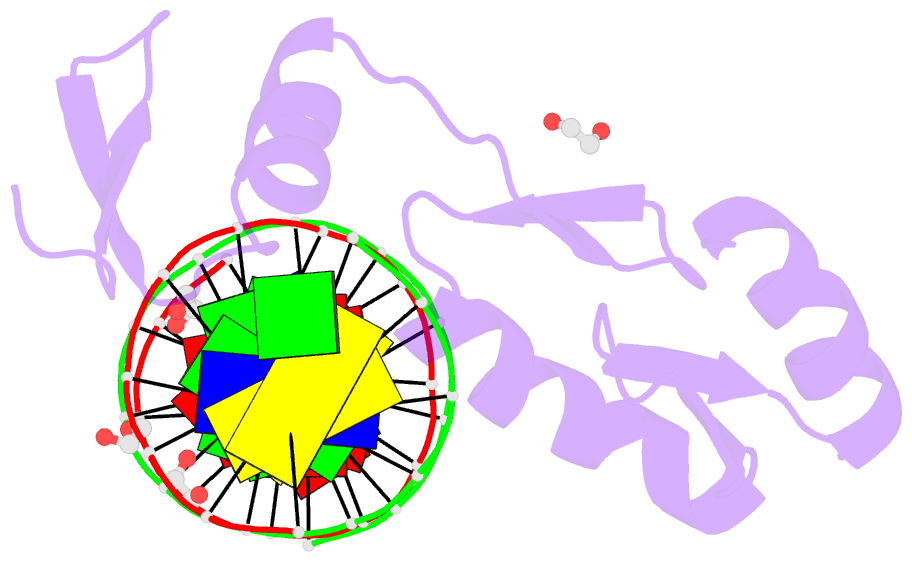
Summary information and primary citation
- PDB-id
- 7n5w; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.24 Å)
- Summary
- Zbtb7a zinc finger domain bound to DNA duplex containing ggaccc (oligo 23)
- Reference
- Ren R, Horton JR, Chen Q, Yang J, Liu B, Huang Y, Blumenthal RM, Zhang X, Cheng X (2023): "Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia." J.Biol.Chem., 299, 102885. doi: 10.1016/j.jbc.2023.102885.
- Abstract
- ZBTB7A belongs to a small family of transcription factors having three members in humans (7A, 7B, and 7C). They share a BTB/POZ protein interaction domain at the amino end and a zinc-finger DNA-binding domain at the carboxyl end. They control the transcription of a wide range of genes, having varied functions in hematopoiesis, oncogenesis, and metabolism (in particular glycolysis). ZBTB7A binding profiles at gene promoters contain a consensus G(a/c)CCC motif, followed by a CCCC sequence in some instances. Structural and mutational investigations suggest that DNA-specific contacts with the four-finger tandem array of ZBTB7A are formed sequentially, initiated from ZF1-ZF2 binding to G(a/c)CCC before spreading to ZF3-ZF4, which bind the DNA backbone and the 3' CCCC sequence respectively. Here we studied some mutations found in t(8;21)-positive acute myeloid leukemia patients that occur within the ZBTB7A DNA-binding domain. We determined that these mutations generally impair ZBTB7A DNA binding, with the most severe disruptions resulting from mutations in ZF1 and ZF2, and the least from a frameshift mutation in ZF3 that results in partial mislocalization. Information provided here on ZBTB7A-DNA interactions is likely applicable to ZBTB7B/C, which have overlapping functions with ZBTB7A in controlling primary metabolism.