Summary information and primary citation
- PDB-id
- 7n8n; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- cryo-EM (3.89 Å)
- Summary
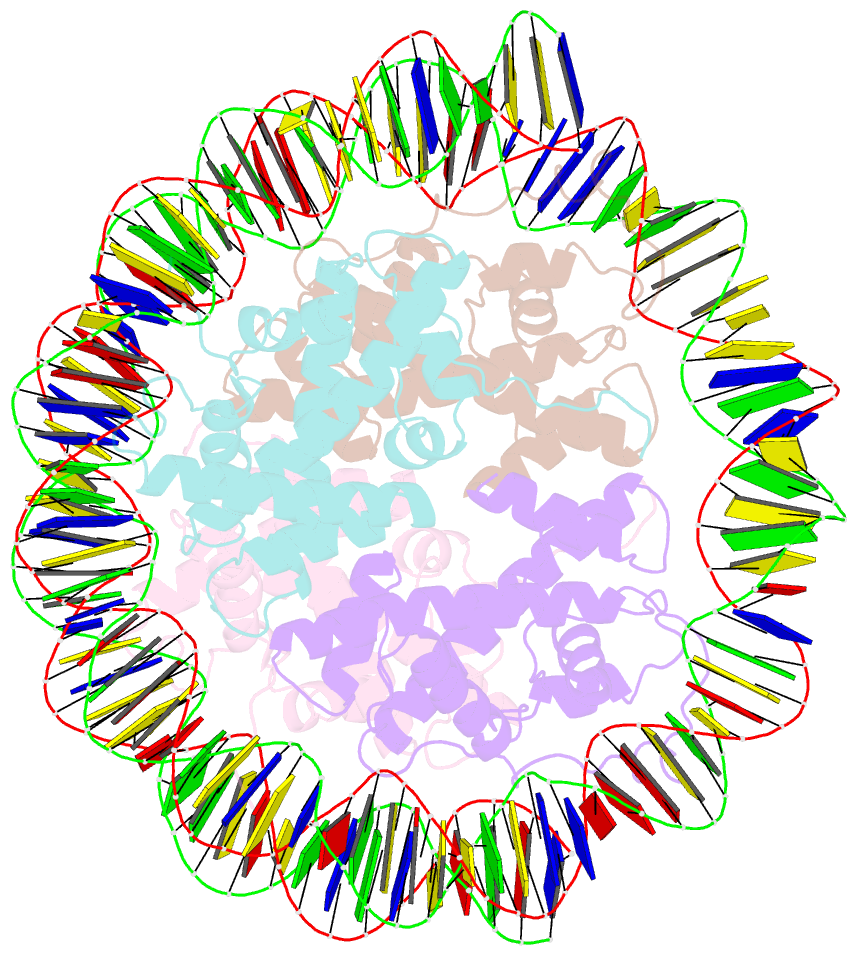
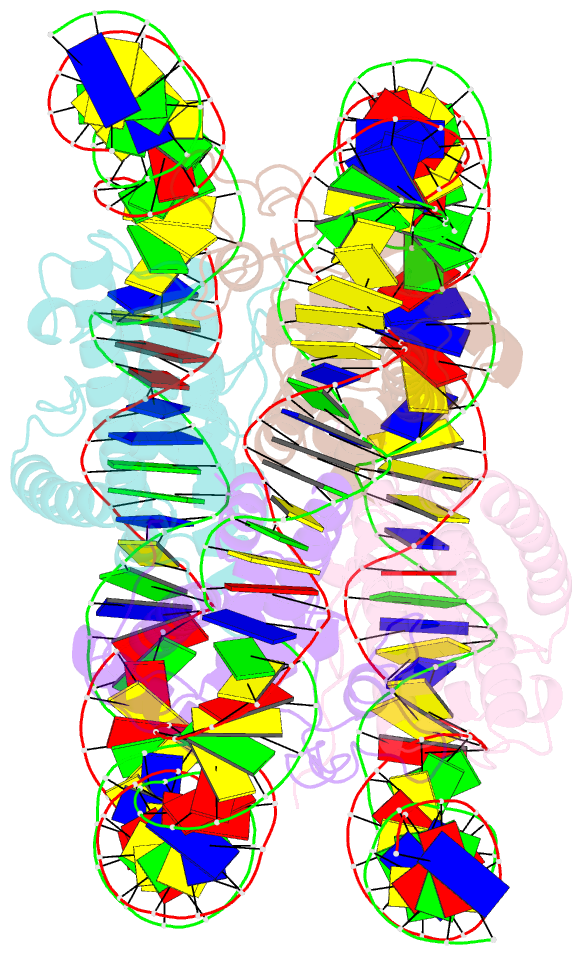
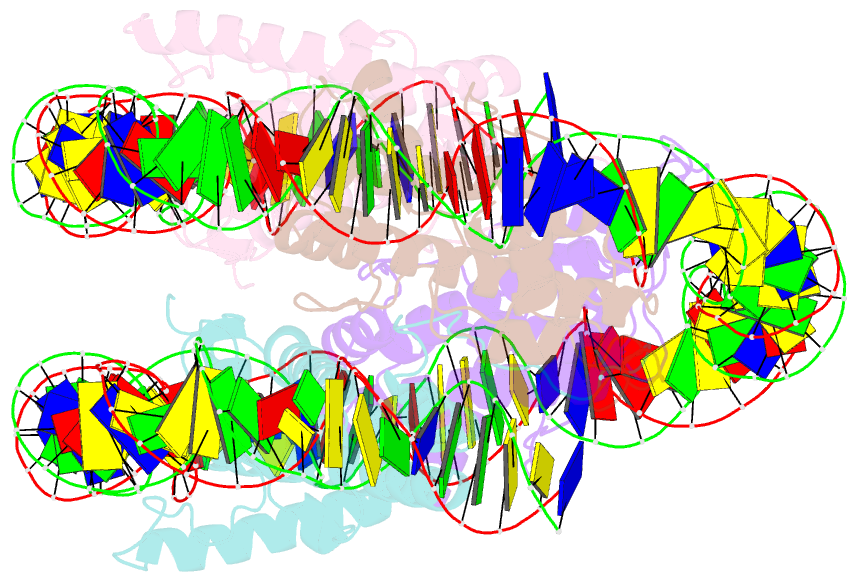
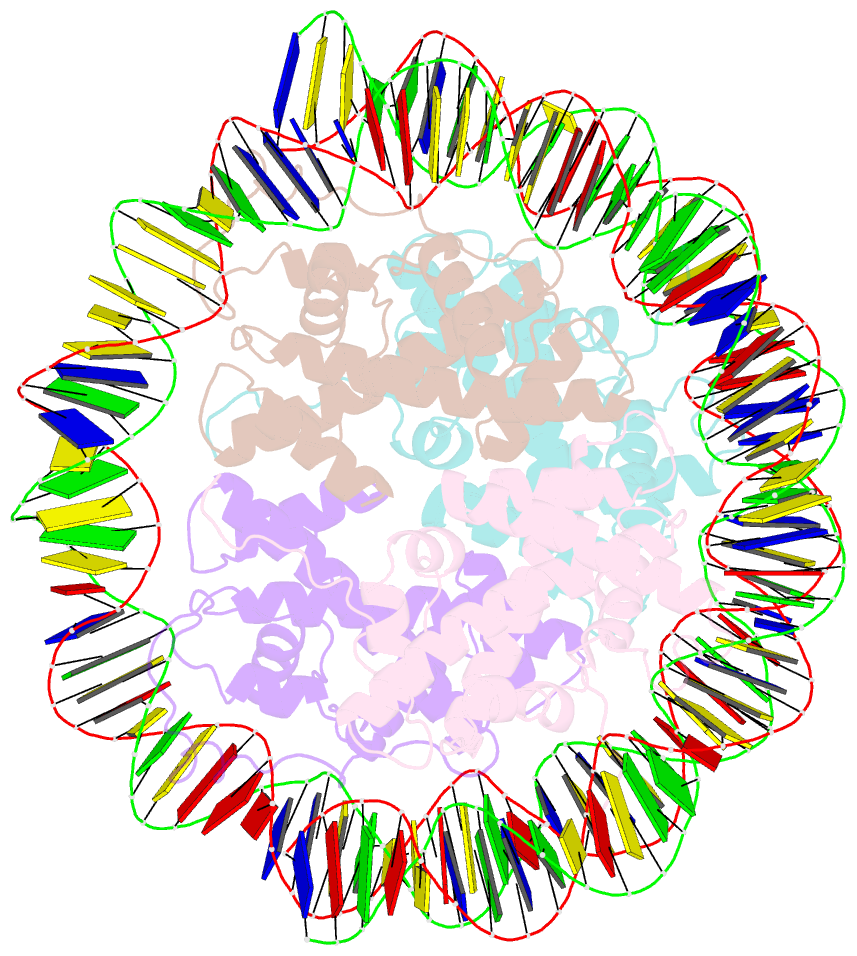
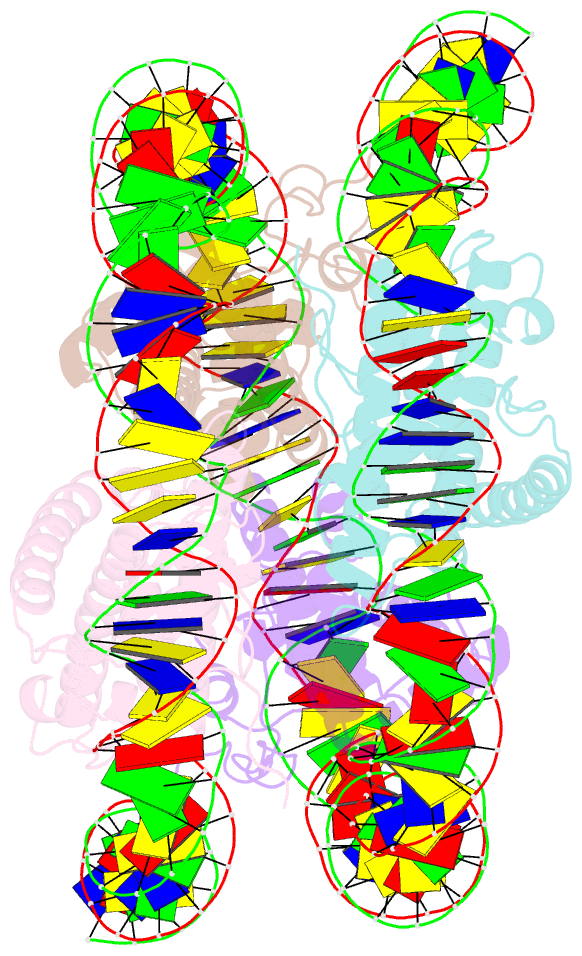
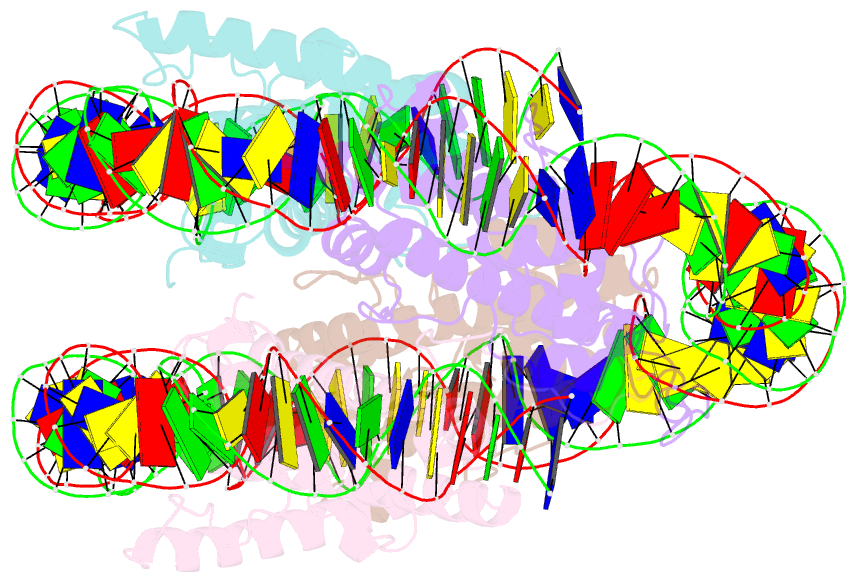
- Melbournevirus nucleosome like particle
- Reference
- Liu Y, Bisio H, Toner CM, Jeudy S, Philippe N, Zhou K, Bowerman S, White A, Edwards G, Abergel C, Luger K (2021): "Virus-encoded histone doublets are essential and form nucleosome-like structures." Cell, 184, 4237-4250.e19. doi: 10.1016/j.cell.2021.06.032.
- Abstract
- The organization of genomic DNA into defined nucleosomes has long been viewed as a hallmark of eukaryotes. This paradigm has been challenged by the identification of "minimalist" histones in archaea and more recently by the discovery of genes that encode fused remote homologs of the four eukaryotic histones in Marseilleviridae, a subfamily of giant viruses that infect amoebae. We demonstrate that viral doublet histones are essential for viral infectivity, localize to cytoplasmic viral factories after virus infection, and ultimately are found in the mature virions. Cryogenic electron microscopy (cryo-EM) structures of viral nucleosome-like particles show strong similarities to eukaryotic nucleosomes despite the limited sequence identify. The unique connectors that link the histone chains contribute to the observed instability of viral nucleosomes, and some histone tails assume structural roles. Our results further expand the range of "organisms" that require nucleosomes and suggest a specialized function of histones in the biology of these unusual viruses.