Summary information and primary citation
- PDB-id
- 7ndj; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein
- Method
- X-ray (1.649 Å)
- Summary
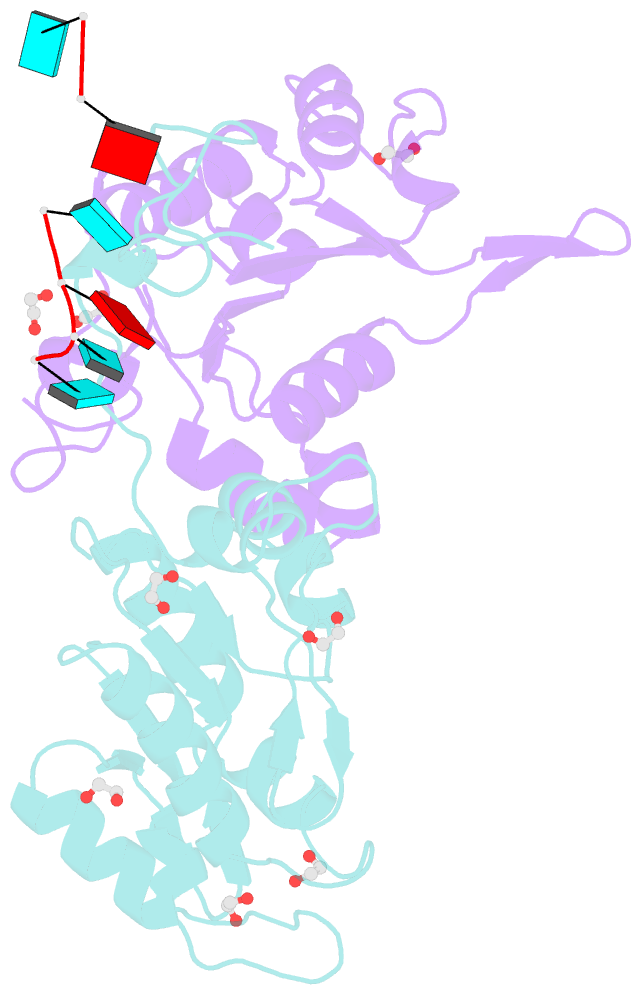
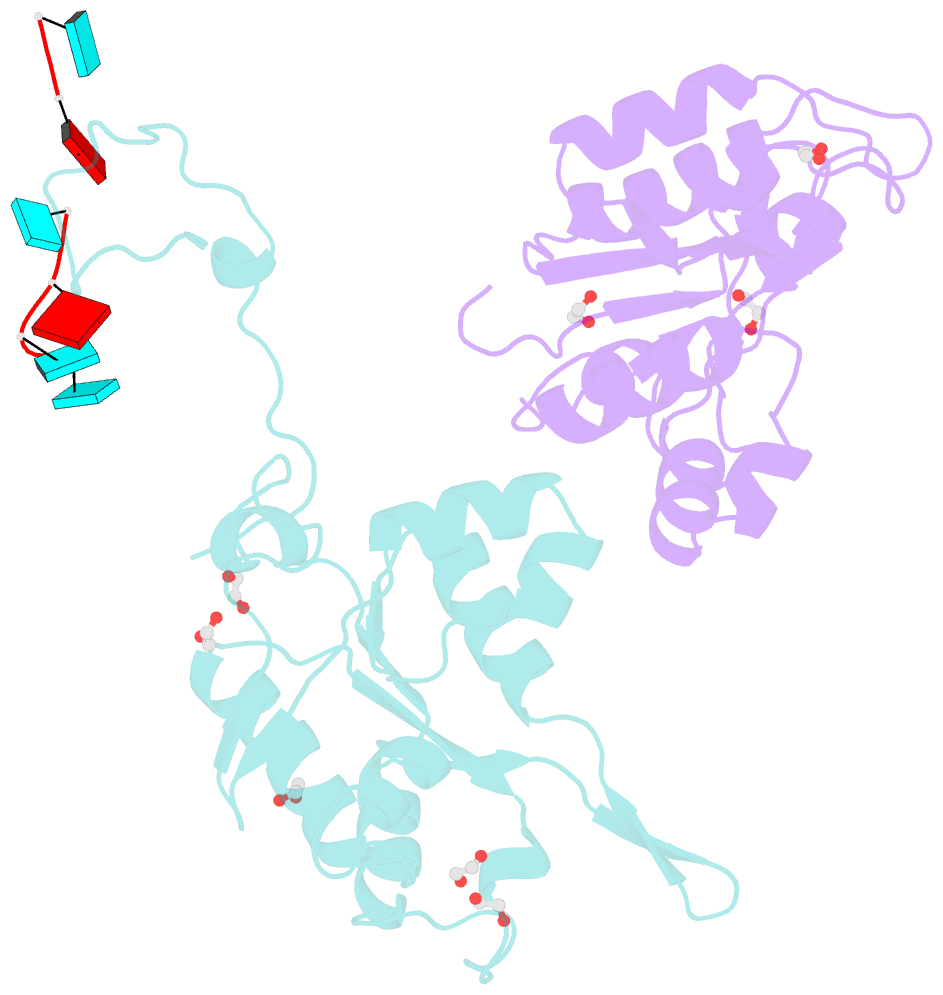
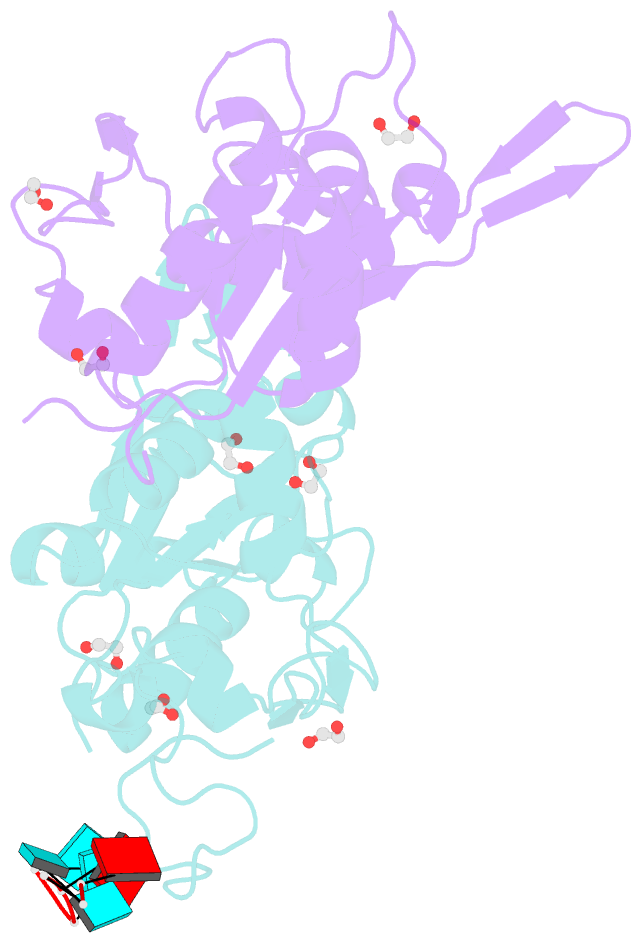
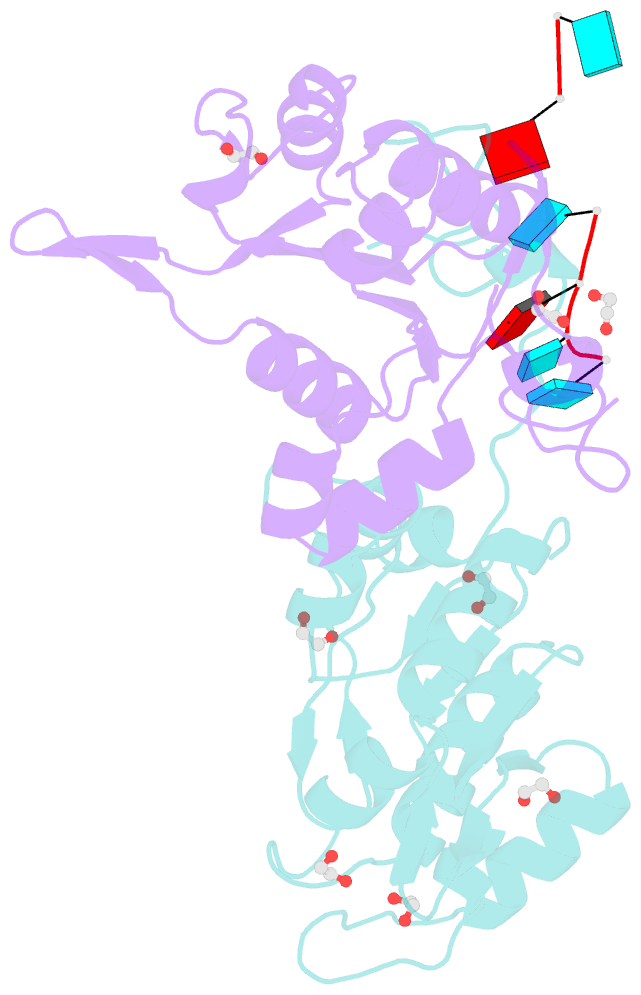
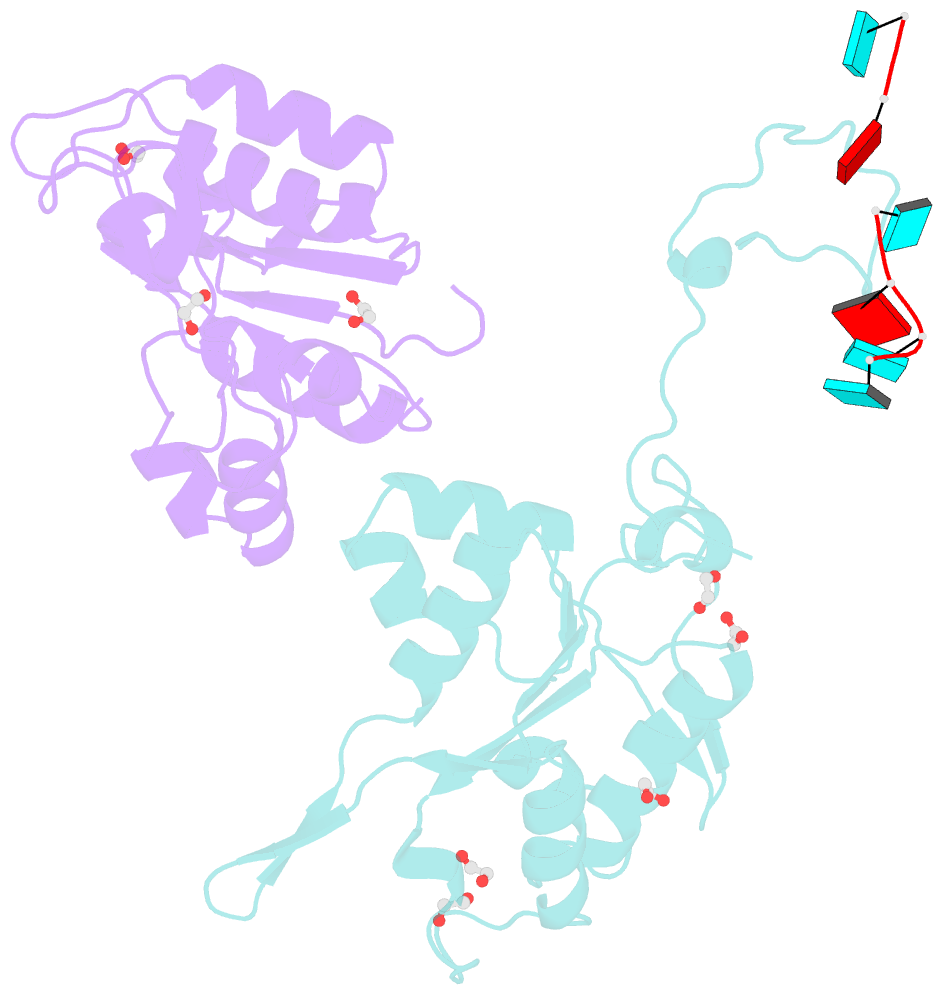
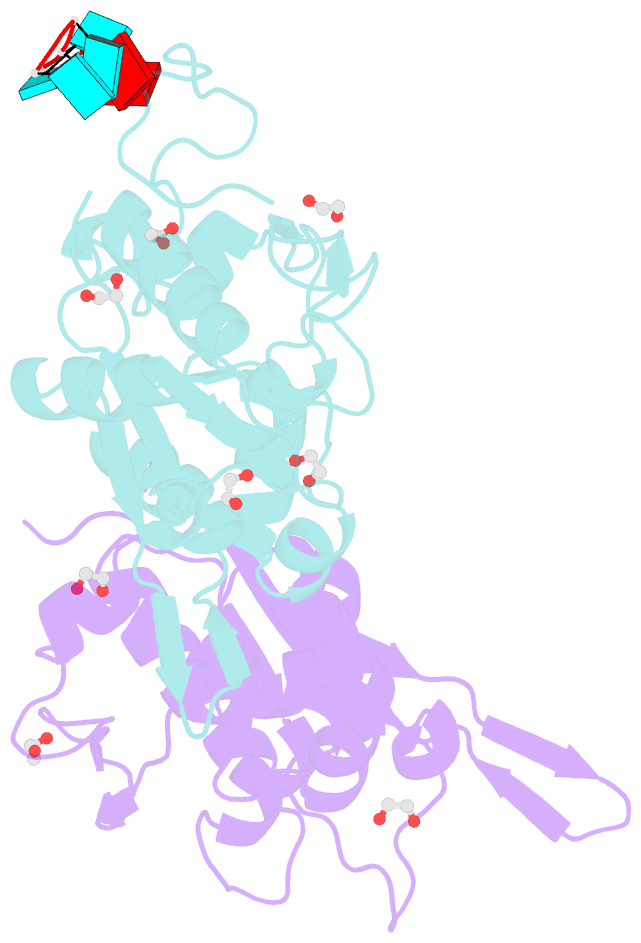
- Crystal structure of zc3h12c pin-ccch zn finger domain with RNA heptamer
- Reference
- Garg A, Roske Y, Yamada S, Uehata T, Takeuchi O, Heinemann U (2021): "PIN and CCCH Zn-finger domains coordinate RNA targeting in ZC3H12 family endoribonucleases." Nucleic Acids Res., 49, 5369-5381. doi: 10.1093/nar/gkab316.
- Abstract
- The CCCH-type zinc finger (ZnF) containing ZC3H12 ribonucleases are crucial in post-transcriptional immune homoeostasis with ZC3H12A being the only structurally studied member of the family. In this study, we present a structural-biochemical characterization of ZC3H12C, which is linked with chronic immune disorders like psoriasis. We established that the RNA substrate is cooperatively recognized by the PIN and ZnF domains of ZC3H12C and analyzed the crystal structure of ZC3H12C bound to a single-stranded RNA substrate. The RNA engages in hydrogen-bonded contacts and stacking interactions with the PIN and ZnF domains simultaneously. The ZC3H12 ZnF shows unprecedented structural features not previously observed in any member of the CCCH-ZnF family and utilizes stacking interactions via a unique combination of spatially conserved aromatic residues to align the target transcript in a bent conformation onto the ZnF scaffold. Further comparative structural analysis of ZC3H12 CCCH-ZnF suggests that a trinucleotide sequence is recognized by ZC3H12 ZnF in target RNA. Our work not only describes the initial structure-biochemical study on ZC3H12C, but also provides the first molecular insight into RNA recognition by a ZC3H12 family member. Finally, our work points to an evolutionary code for RNA recognition adopted by CCCH-type ZnF proteins.