Summary information and primary citation
- PDB-id
- 7nfe; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- cryo-EM (4.29 Å)
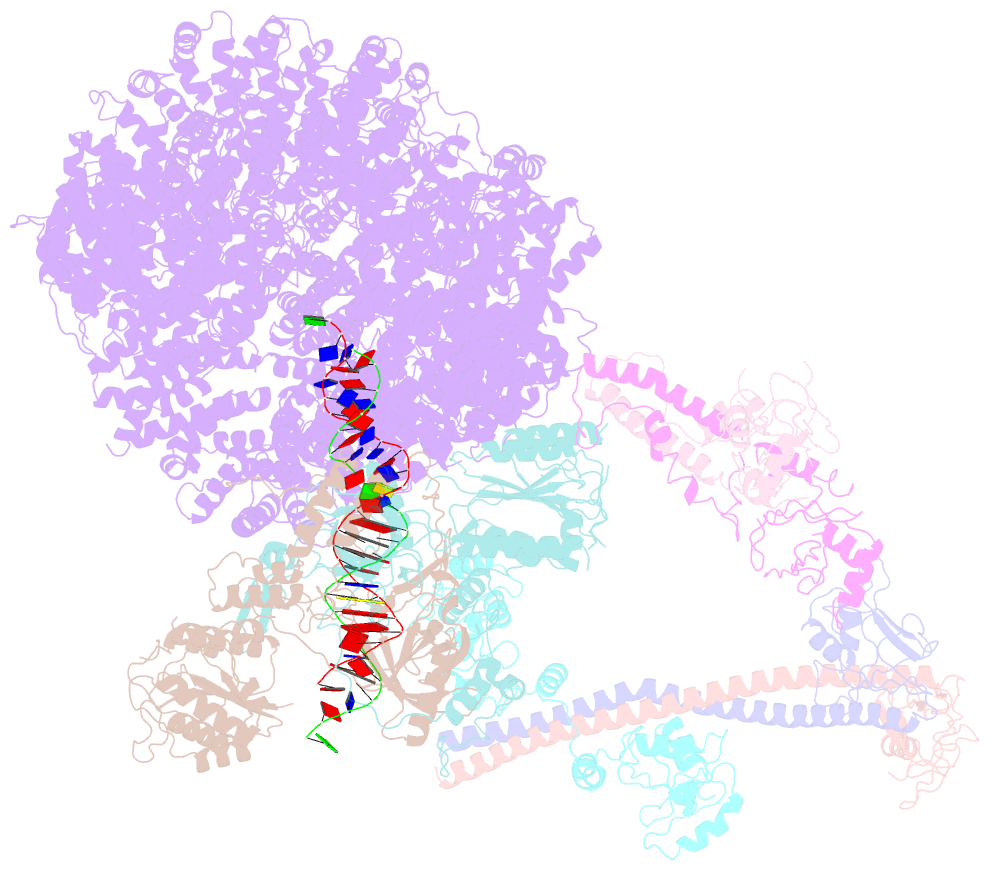
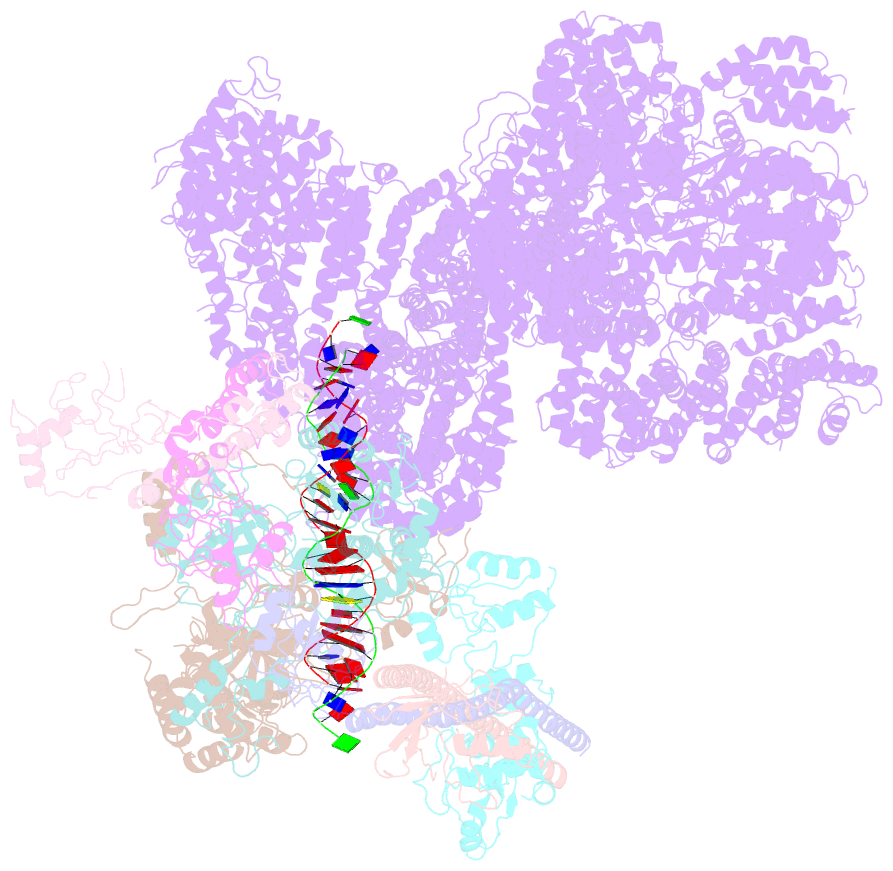
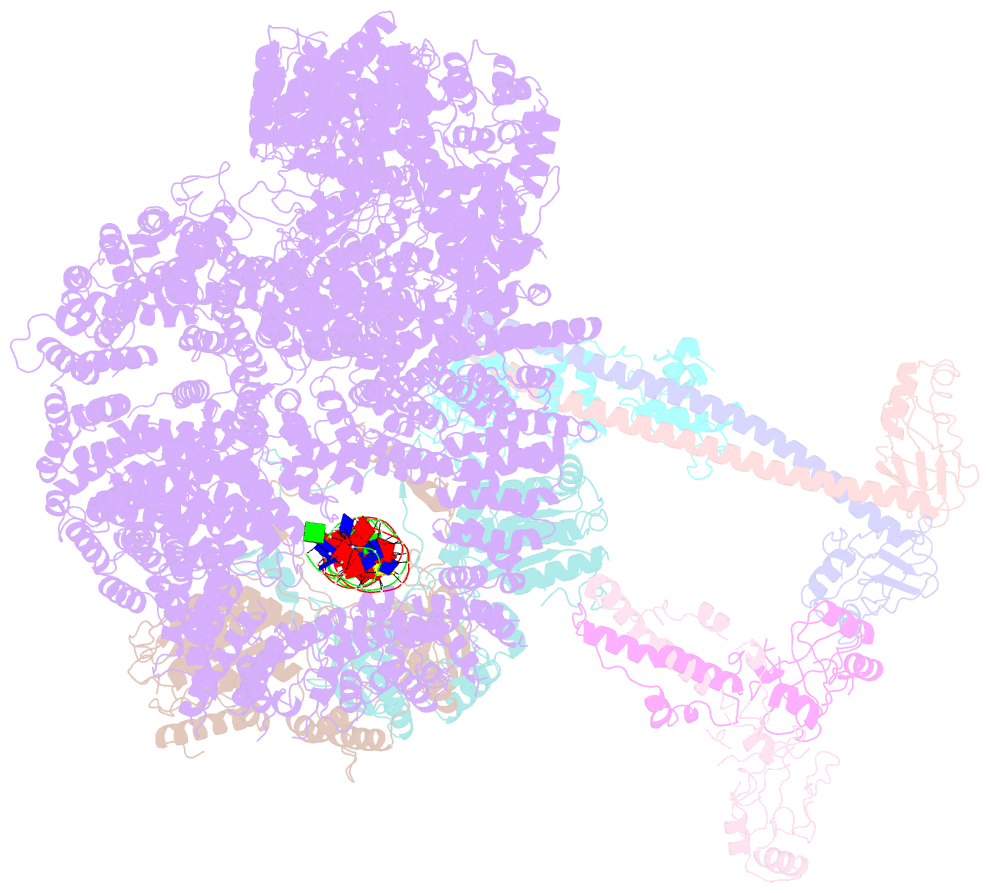
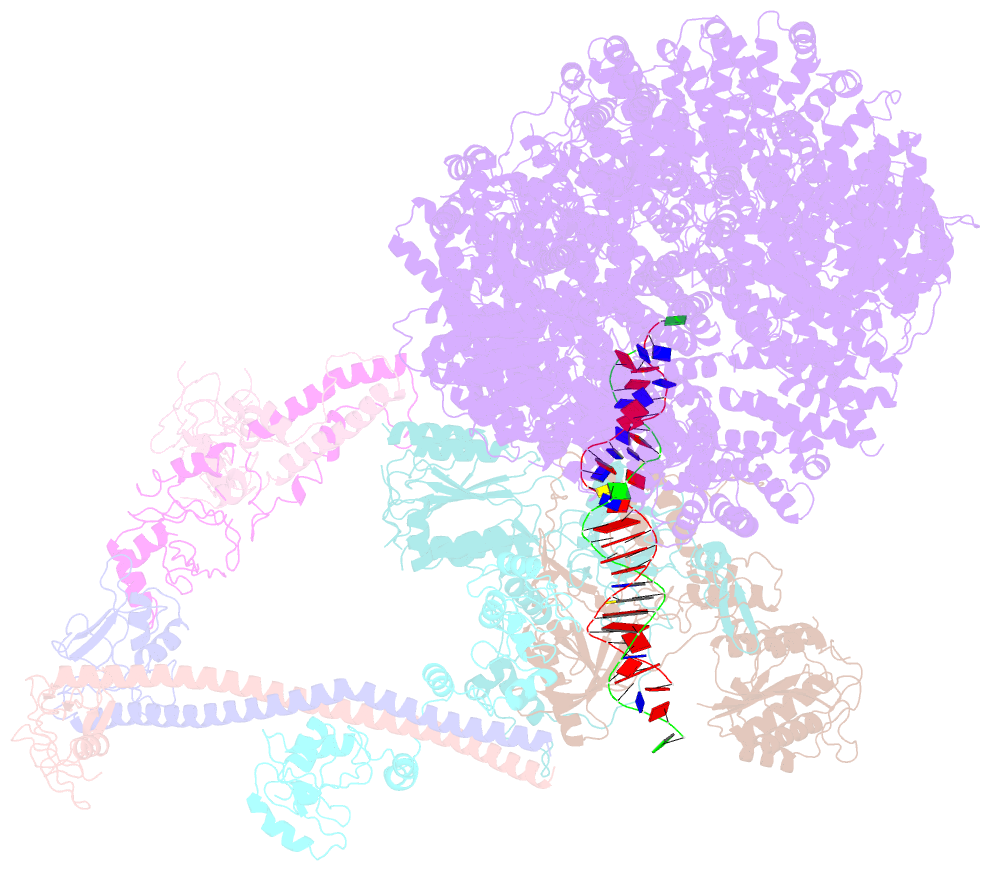
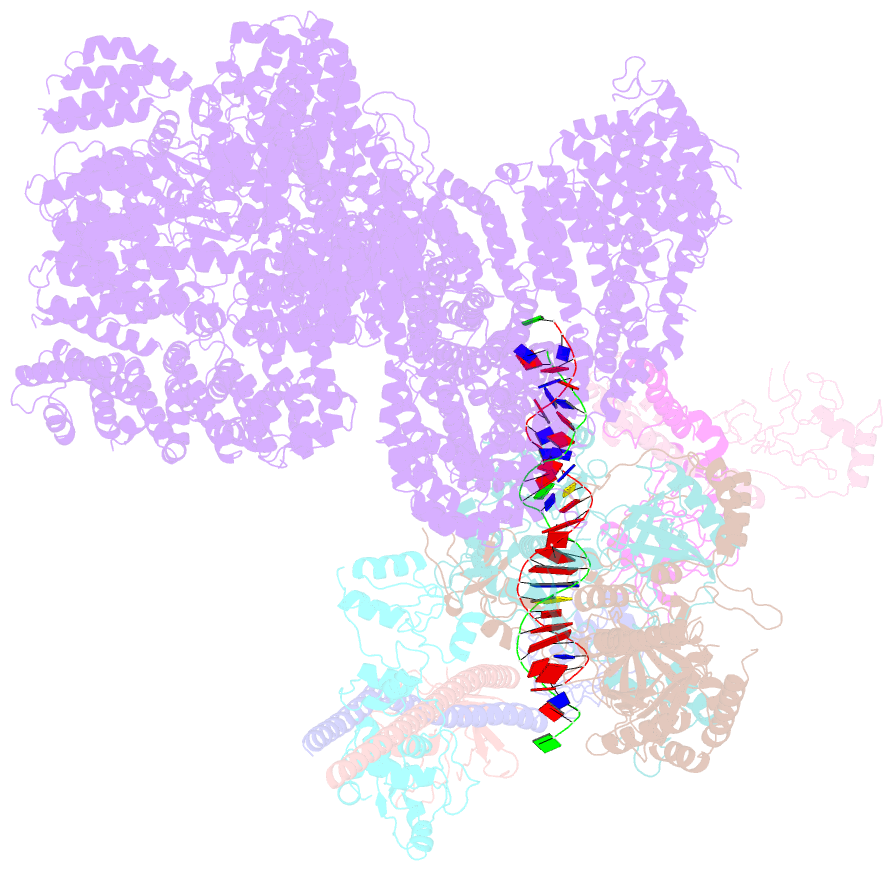
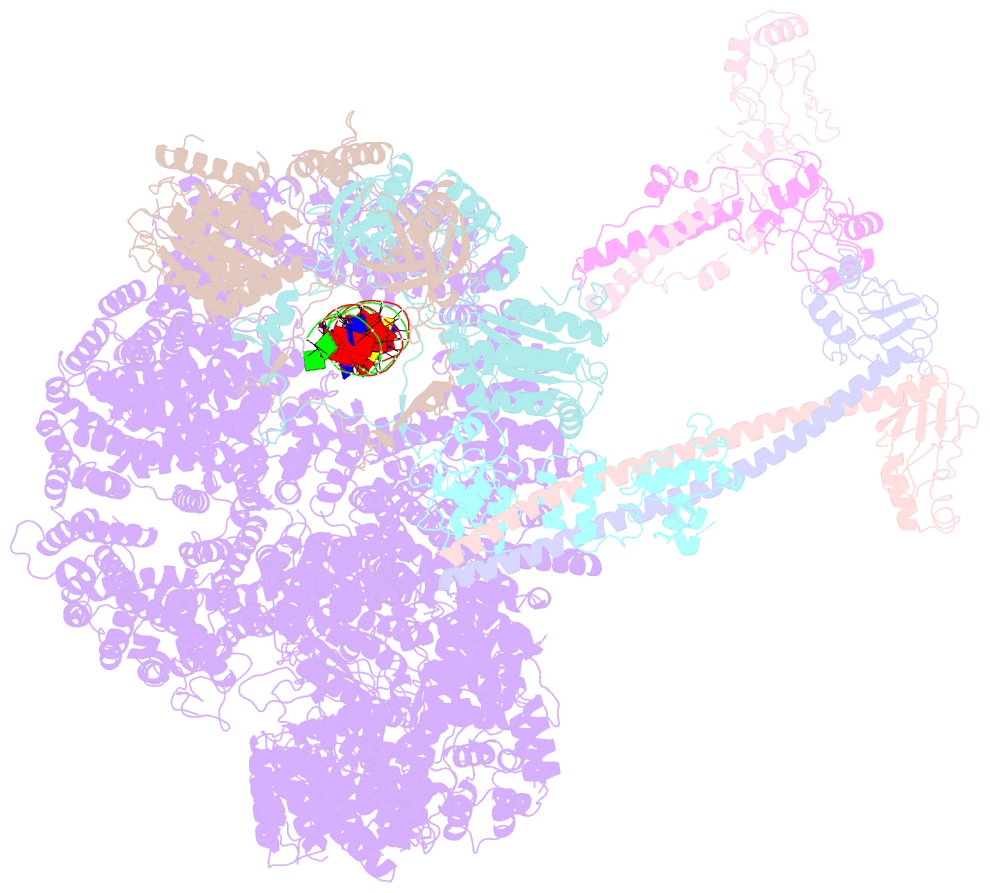
- Summary
- cryo-EM structure of nhej super-complex (monomer)
- Reference
- Chaplin AK, Hardwick SW, Stavridi AK, Buehl CJ, Goff NJ, Ropars V, Liang S, De Oliveira TM, Chirgadze DY, Meek K, Charbonnier JB, Blundell TL (2021): "Cryo-EM of NHEJ supercomplexes provides insights into DNA repair." Mol.Cell, 81, 3400-3409. doi: 10.1016/j.molcel.2021.07.005.
- Abstract
- Non-homologous end joining (NHEJ) is one of two critical mechanisms utilized in humans to repair DNA double-strand breaks (DSBs). Unrepaired or incorrect repair of DSBs can lead to apoptosis or cancer. NHEJ involves several proteins, including the Ku70/80 heterodimer, DNA-dependent protein kinase catalytic subunit (DNA-PKcs), X-ray cross-complementing protein 4 (XRCC4), XRCC4-like factor (XLF), and ligase IV. These core proteins bind DSBs and ligate the damaged DNA ends. However, details of the structural assembly of these proteins remain unclear. Here, we present cryo-EM structures of NHEJ supercomplexes that are composed of these core proteins and DNA, revealing the detailed structural architecture of this assembly. We describe monomeric and dimeric forms of this supercomplex and also propose the existence of alternate dimeric forms of long-range synaptic complexes. Finally, we show that mutational disruption of several structural features within these NHEJ complexes negatively affects DNA repair.