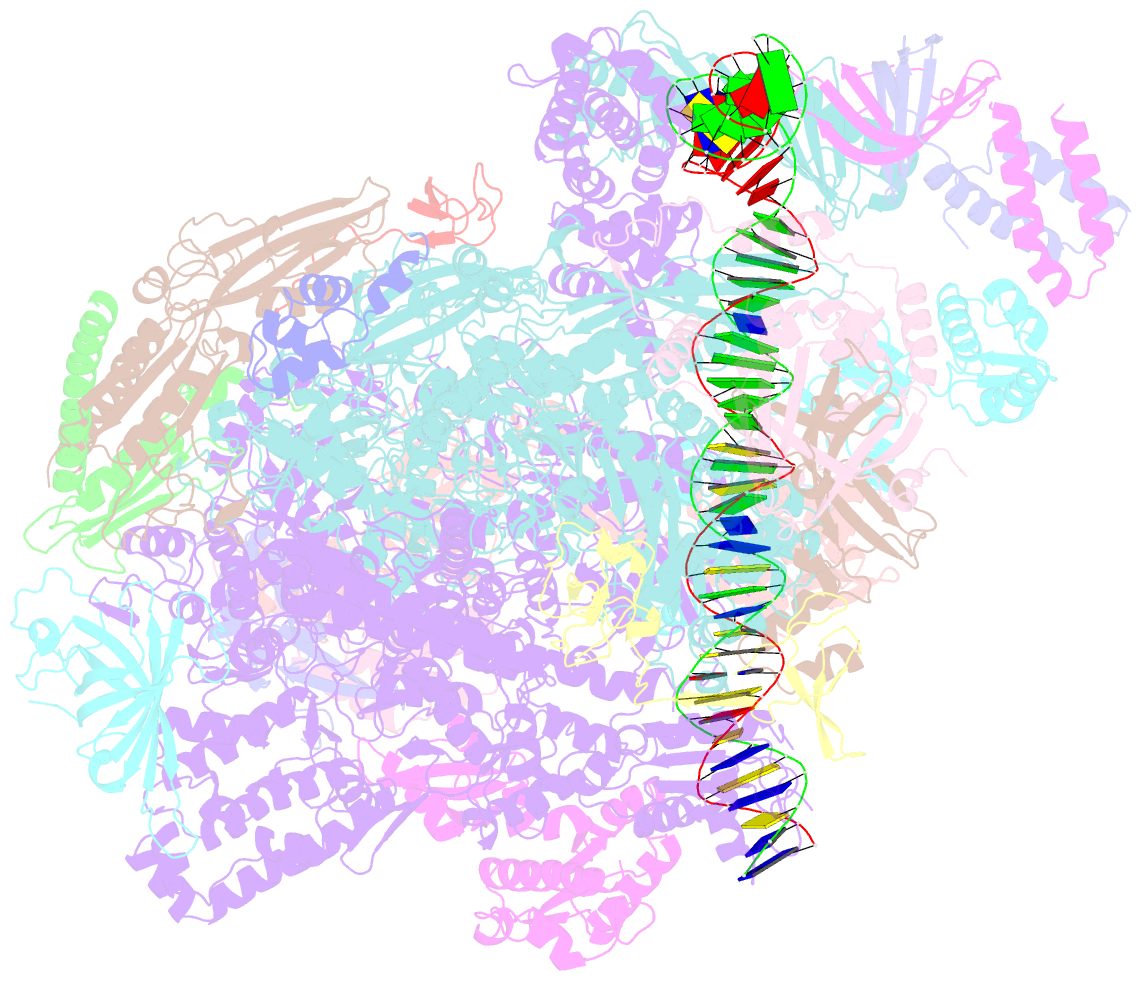
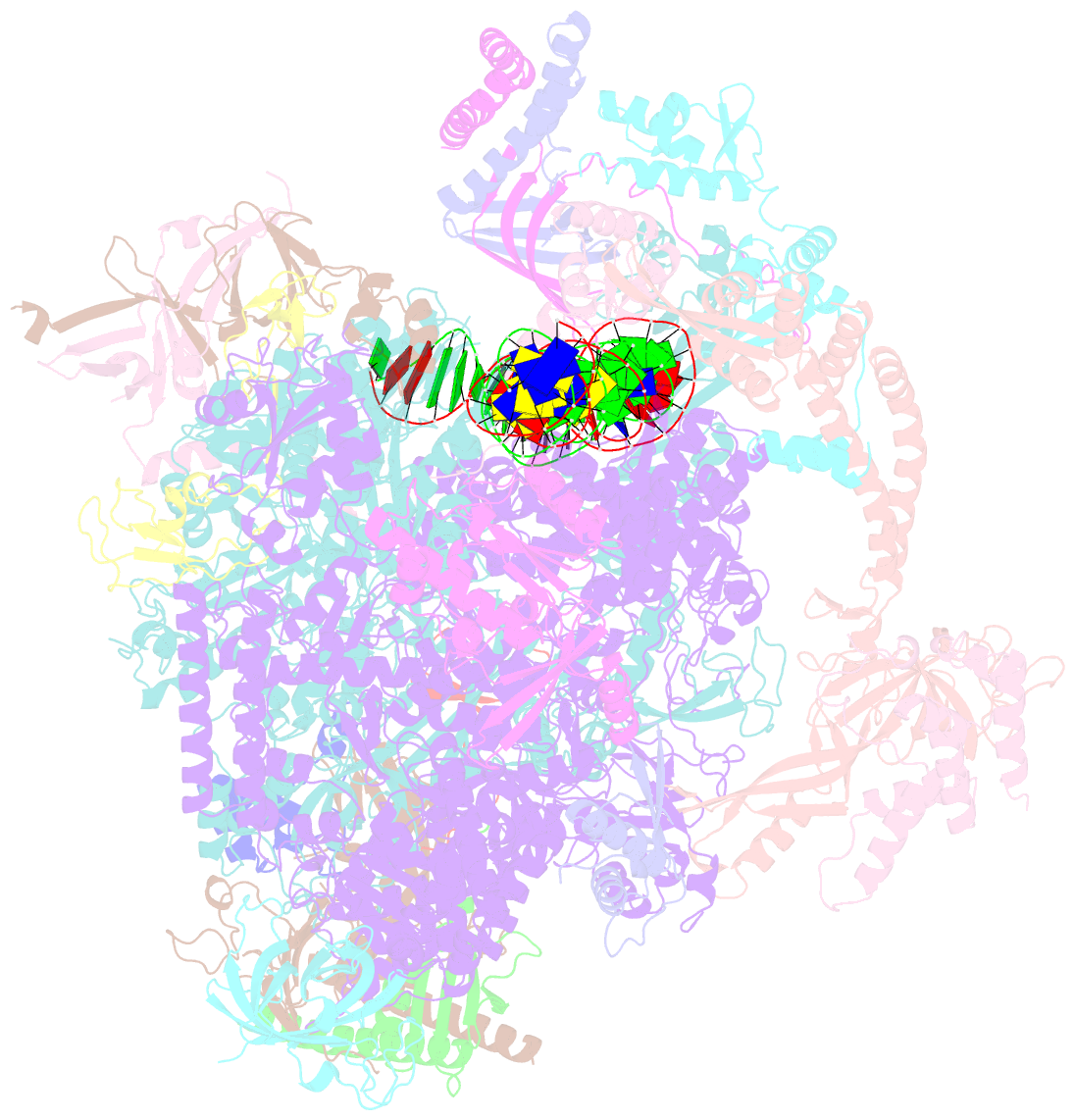
Summary information and primary citation
- PDB-id
- 7nvs; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- cryo-EM (2.8 Å)
- Summary
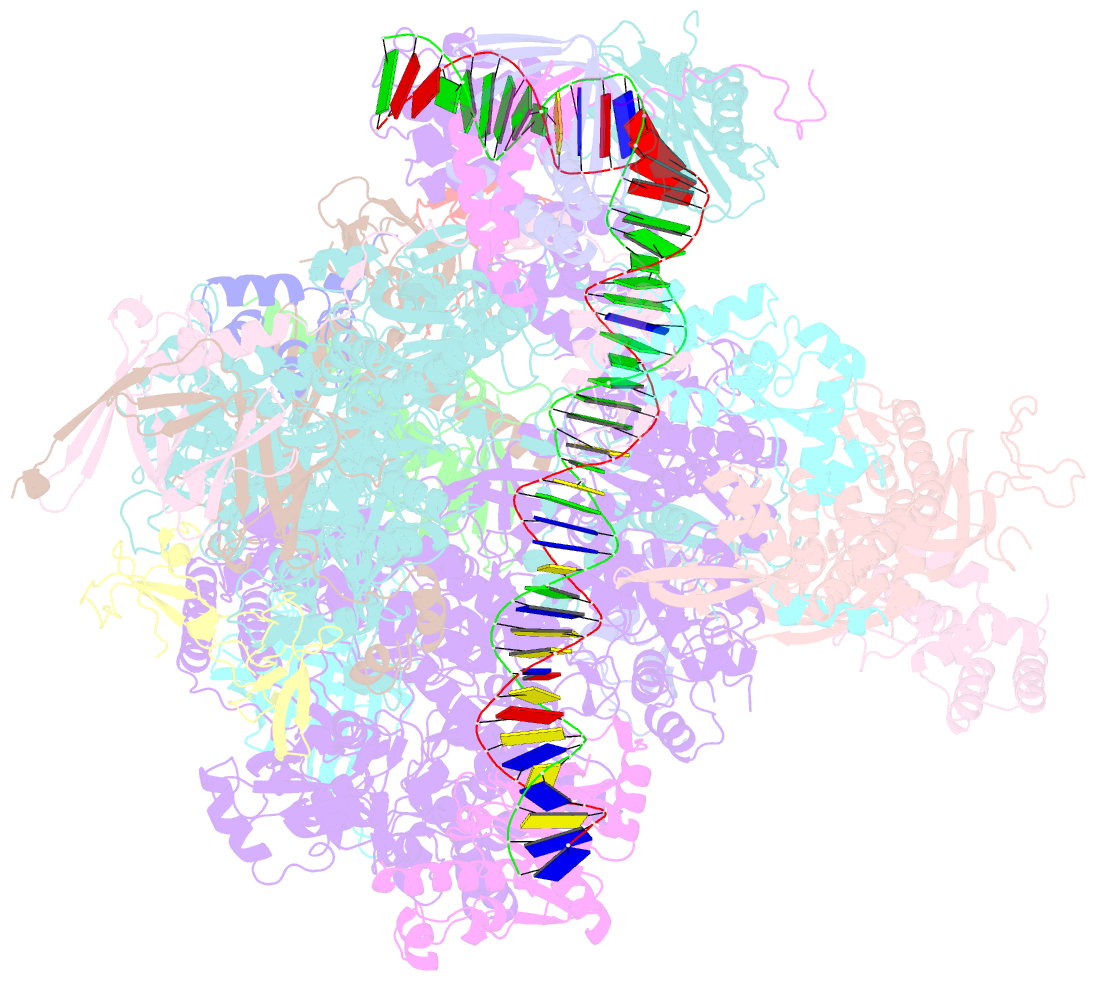
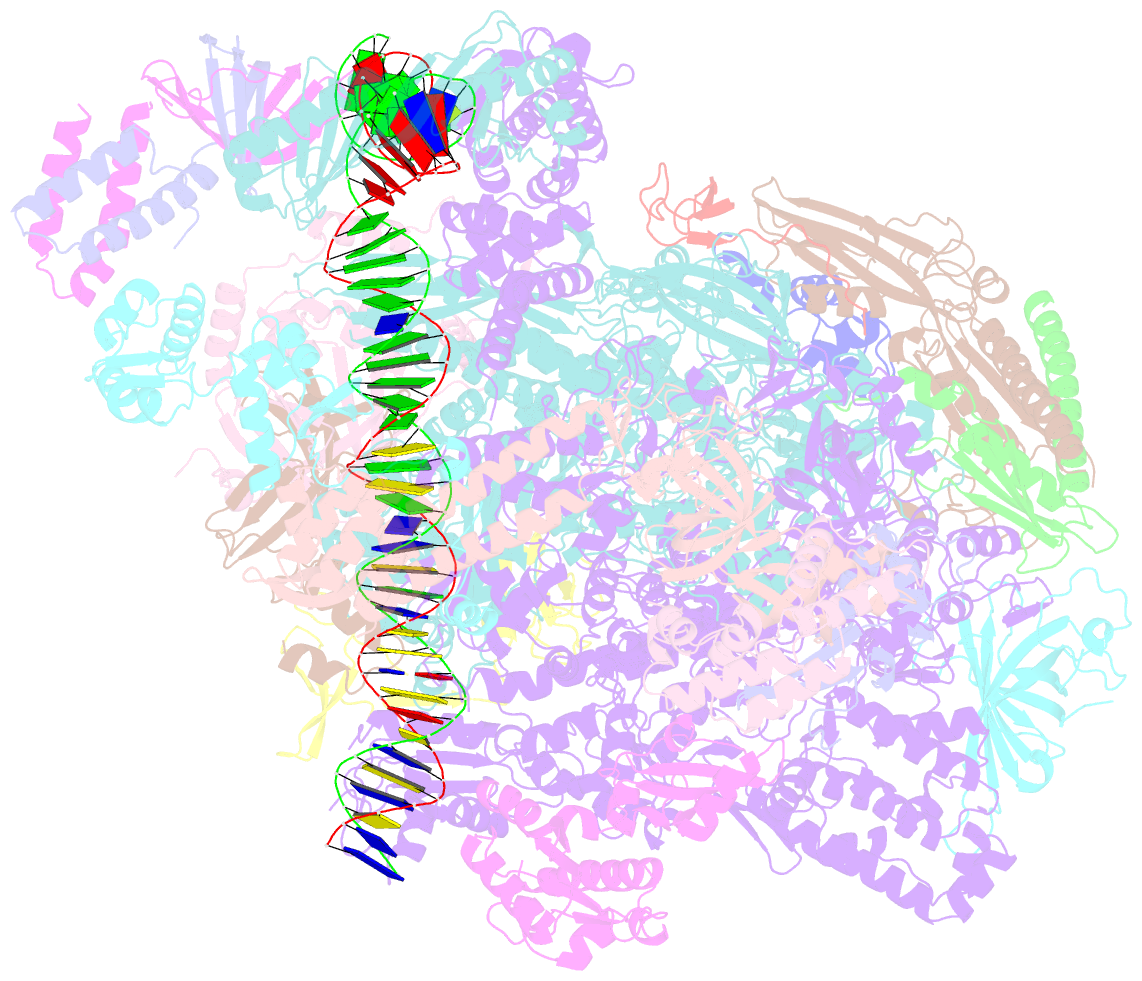
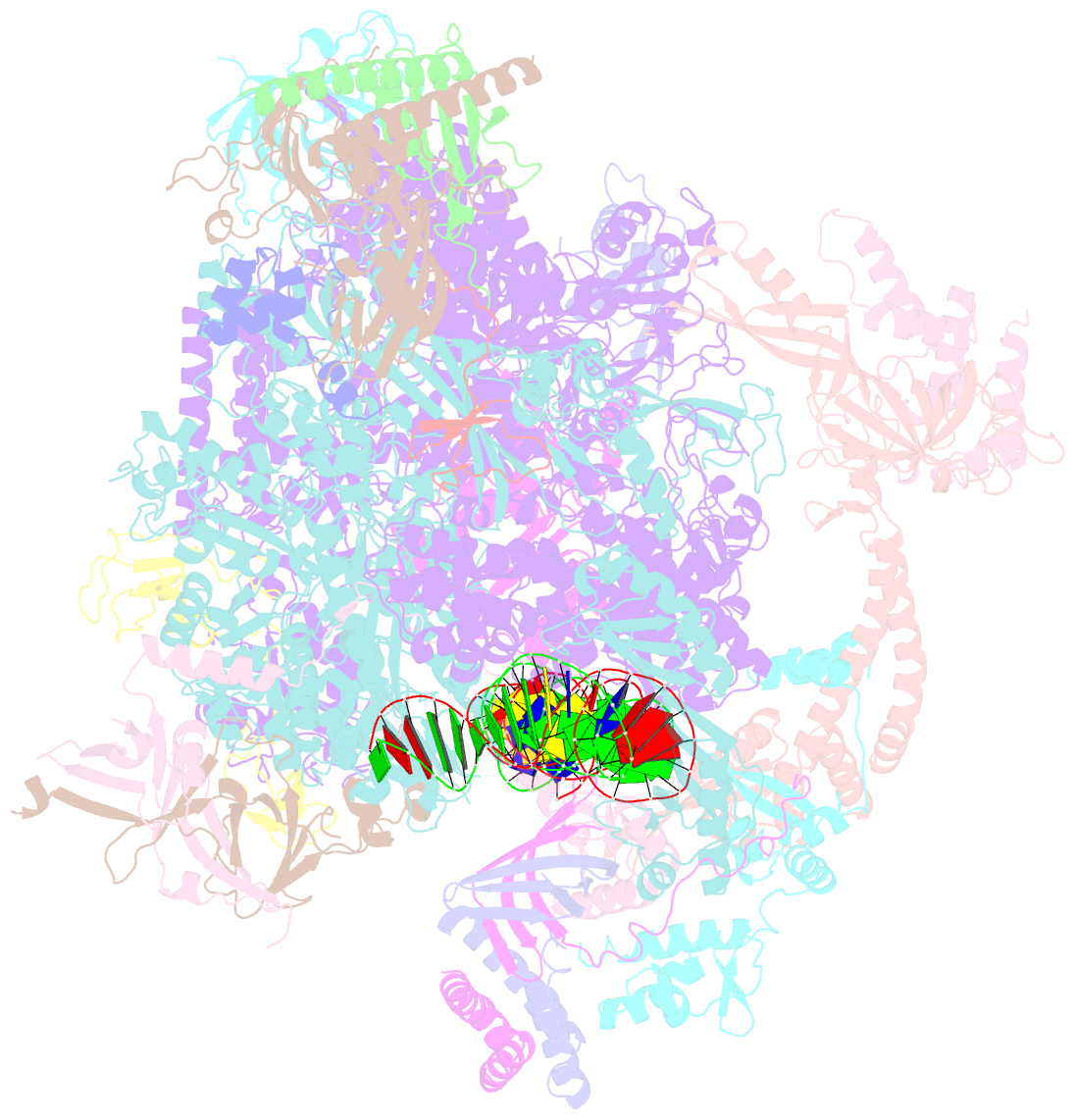
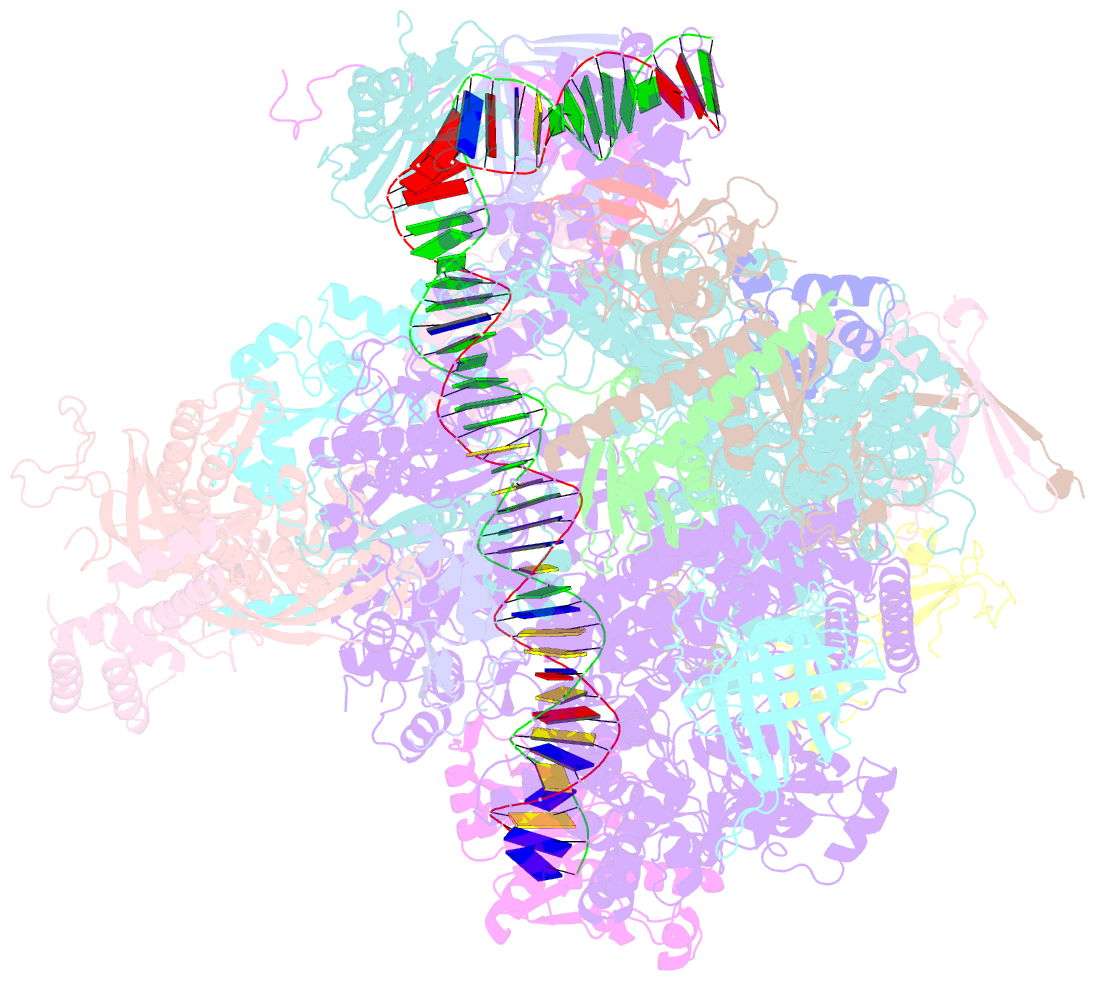
- RNA polymerase ii core pre-initiation complex with closed promoter DNA in proximal position
- Reference
- Aibara S, Schilbach S, Cramer P (2021): "Structures of mammalian RNA polymerase II pre-initiation complexes." Nature, 594, 124-128. doi: 10.1038/s41586-021-03554-8.
- Abstract
- The initiation of transcription is a focal point for the regulation of gene activity during mammalian cell differentiation and development. To initiate transcription, RNA polymerase II (Pol II) assembles with general transcription factors into a pre-initiation complex (PIC) that opens promoter DNA. Previous work provided the molecular architecture of the yeast1-9 and human10,11 PIC and a topological model for DNA opening by the general transcription factor TFIIH12-14. Here we report the high-resolution cryo-electron microscopy structure of PIC comprising human general factors and Sus scrofa domesticus Pol II, which is 99.9% identical to human Pol II. We determine the structures of PIC with closed and opened promoter DNA at 2.5-2.8 Å resolution, and resolve the structure of TFIIH at 2.9-4.0 Å resolution. We capture the TFIIH translocase XPB in the pre- and post-translocation states, and show that XPB induces and propagates a DNA twist to initiate the opening of DNA approximately 30 base pairs downstream of the TATA box. We also provide evidence that DNA opening occurs in two steps and leads to the detachment of TFIIH from the core PIC, which may stop DNA twisting and enable RNA chain initiation.