Summary information and primary citation
- PDB-id
- 7o56; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- X-ray (2.6 Å)
- Summary
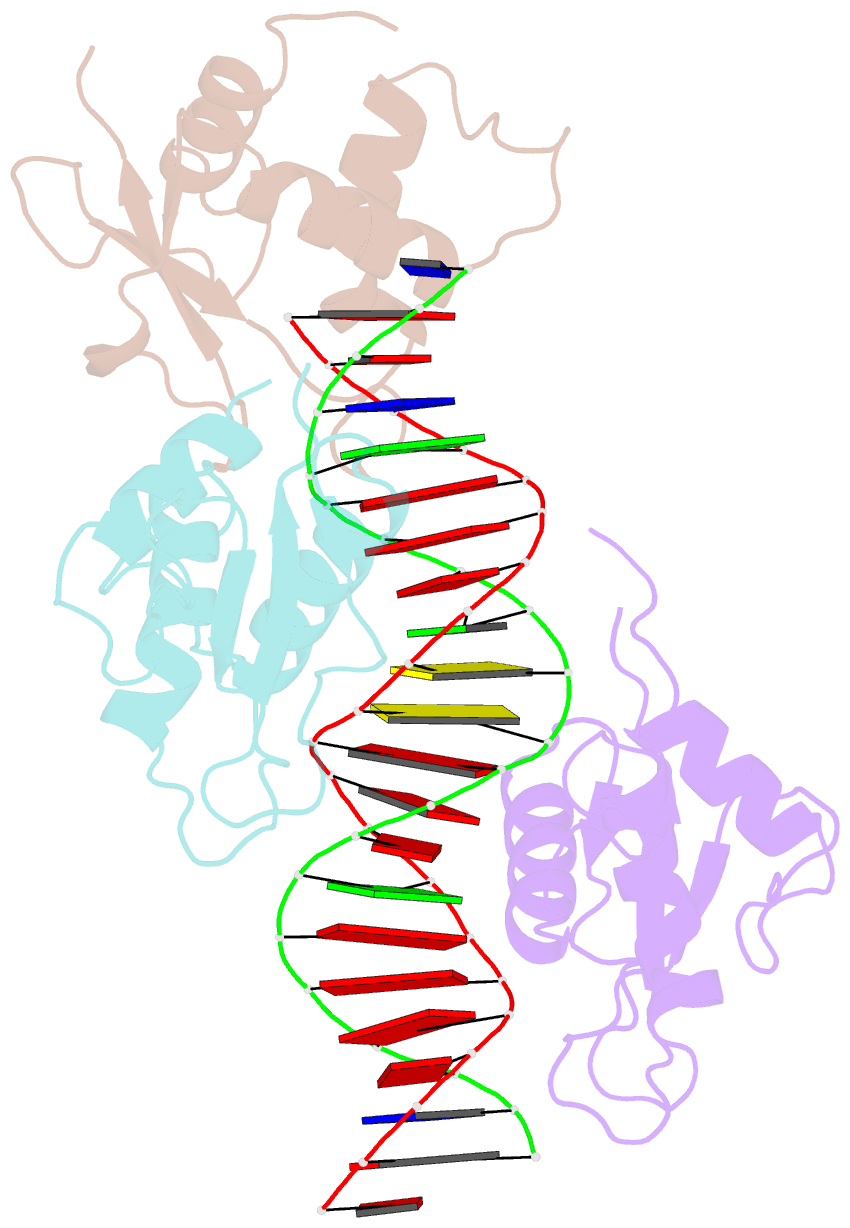
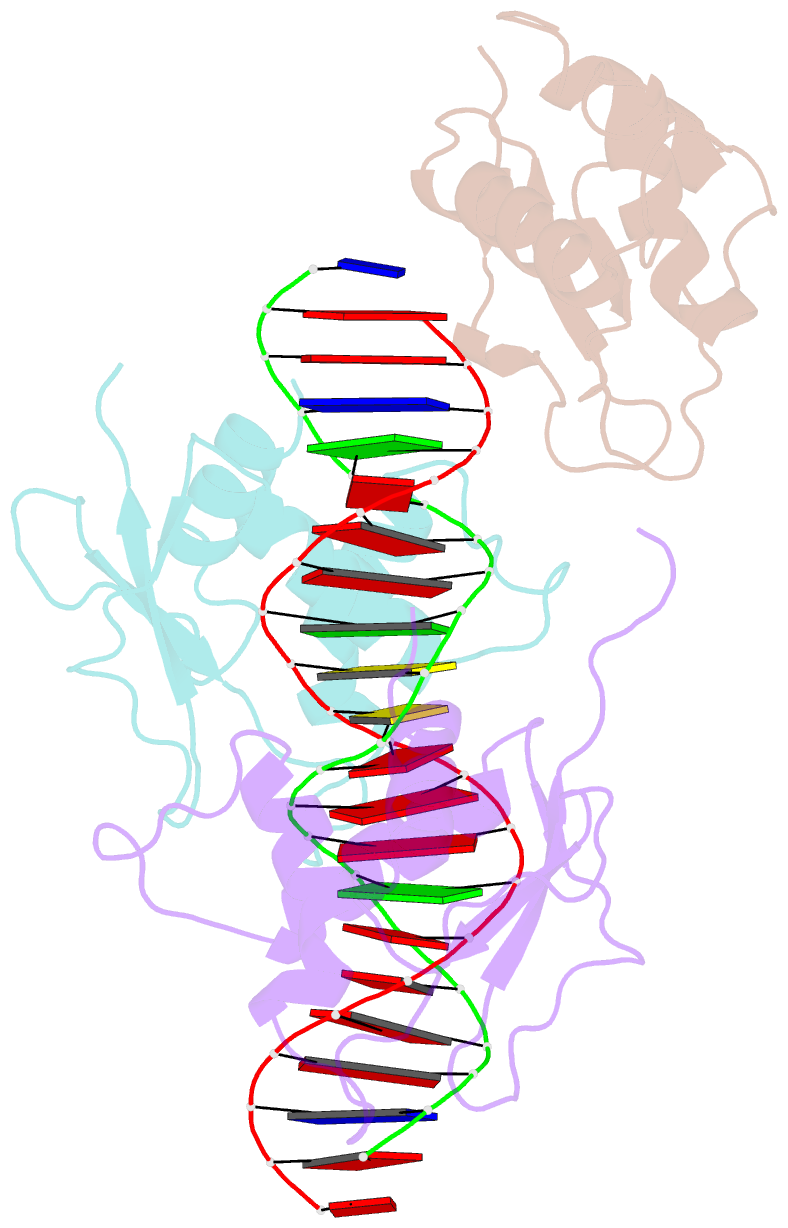
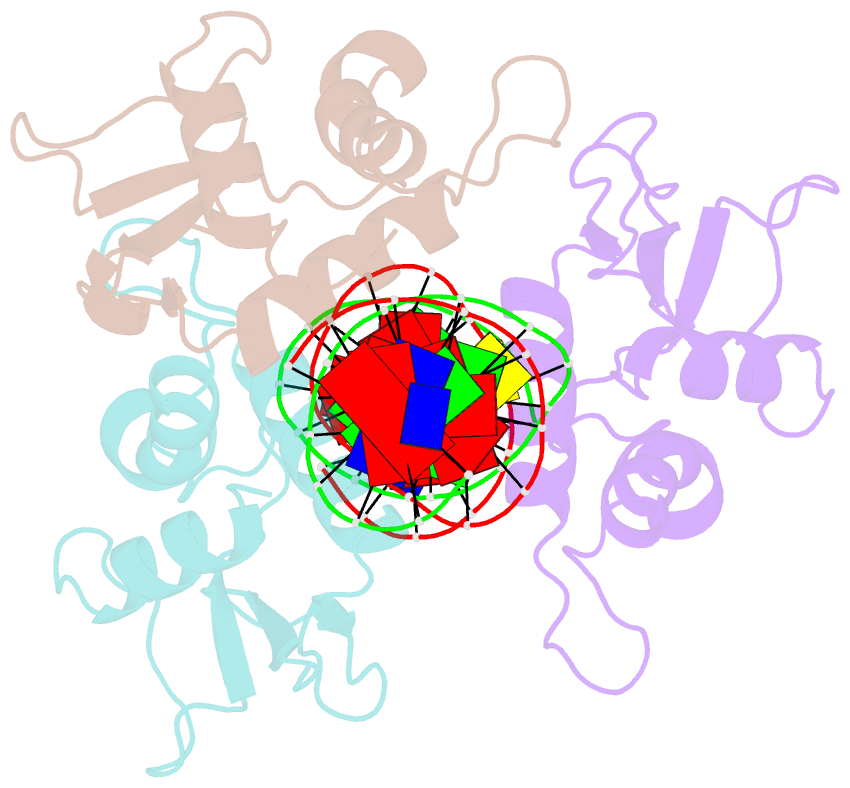
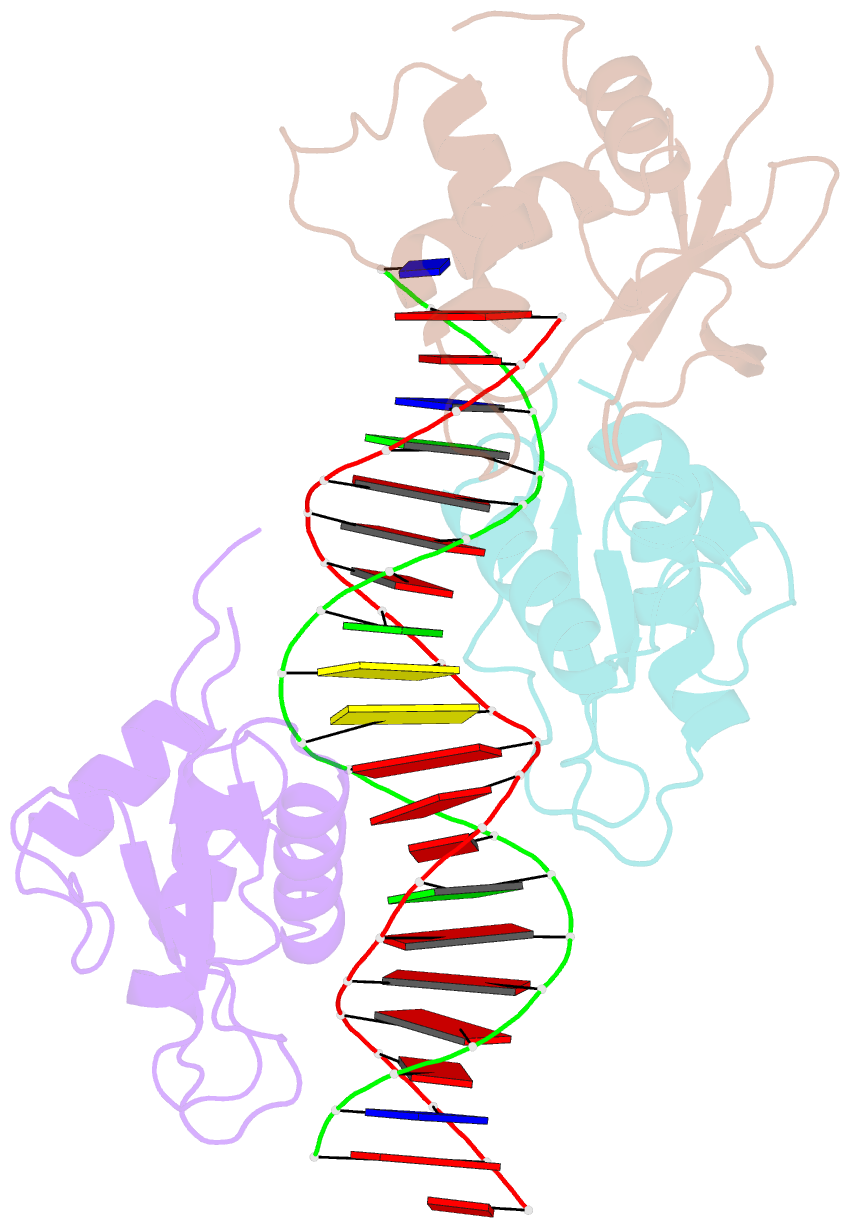
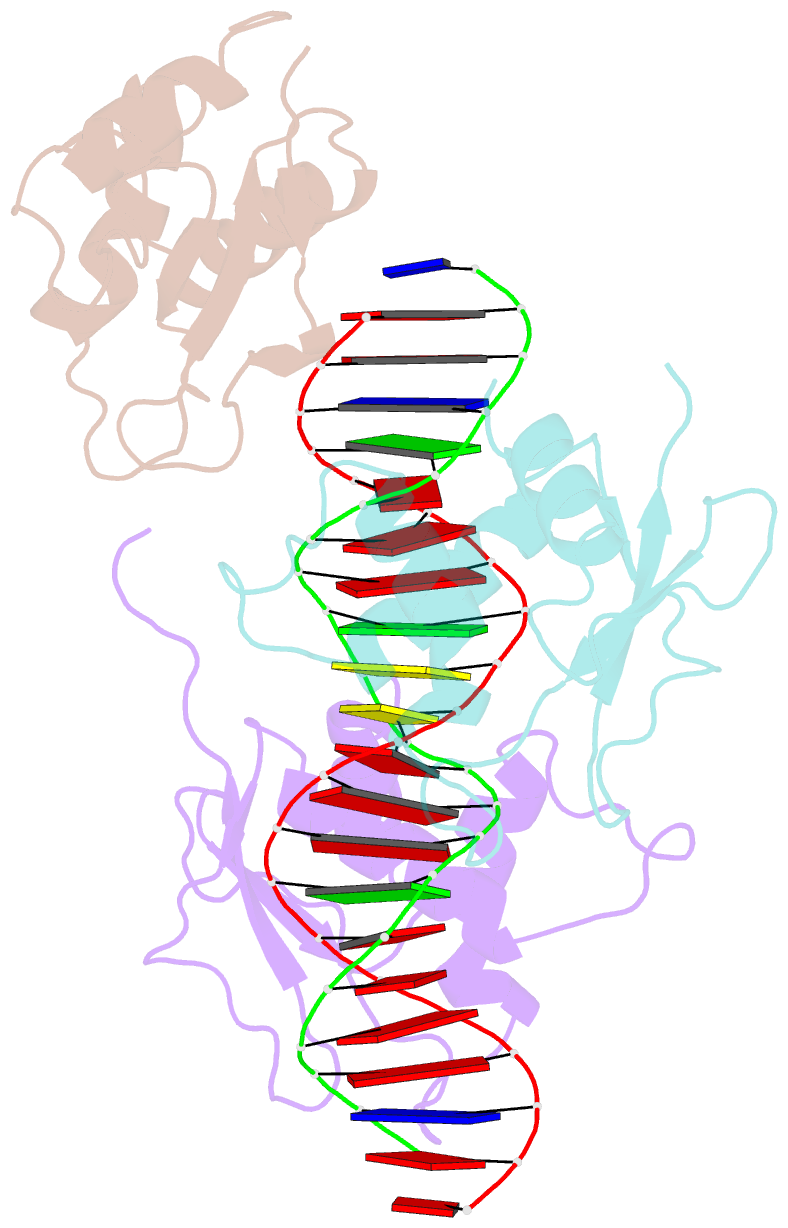
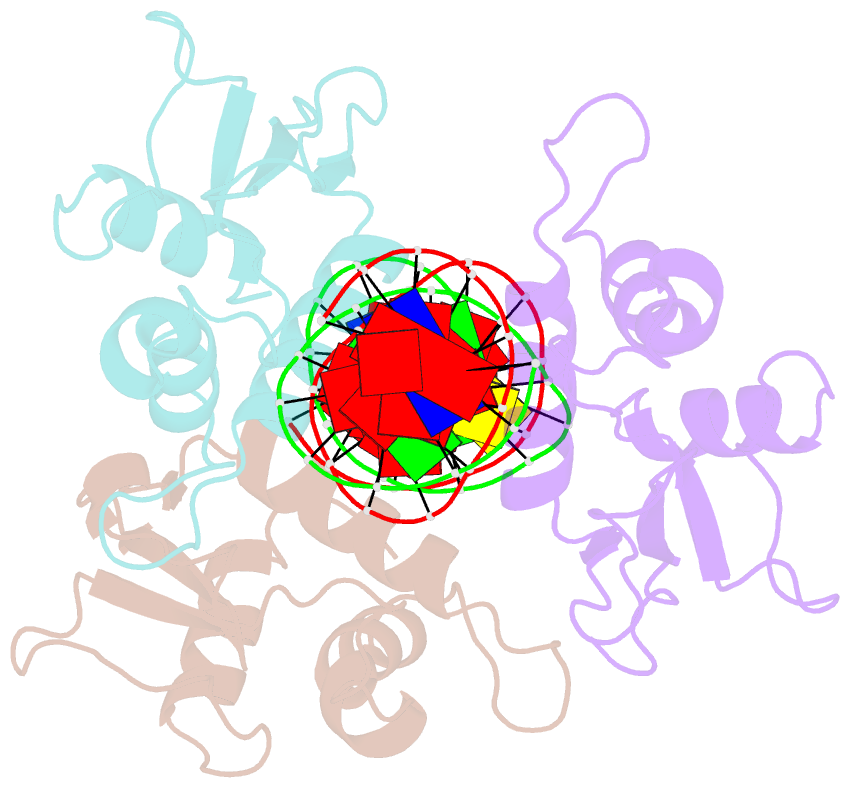
- X-ray structure of interferon regulatory factor 4 DNA binding domain bound to an interferon-stimulated response element solved by phosphorus and sulphur sad methods
- Reference
- Agnarelli A, El Omari K, Duman R, Wagner A, Mancini EJ (2021): "Phosphorus and sulfur SAD phasing of the nucleic acid-bound DNA-binding domain of interferon regulatory factor 4." Acta Crystallogr.,Sect.F, 77, 202-207. doi: 10.1107/S2053230X21006506.
- Abstract
- Pivotal to the regulation of key cellular processes such as the transcription, replication and repair of DNA, DNA-binding proteins play vital roles in all aspects of genetic activity. The determination of high-quality structures of DNA-binding proteins, particularly those in complexes with DNA, provides crucial insights into the understanding of these processes. The presence in such complexes of phosphate-rich oligonucleotides offers the choice of a rapid method for the routine solution of DNA-binding proteins through the use of long-wavelength beamlines such as I23 at Diamond Light Source. This article reports the use of native intrinsic phosphorus and sulfur single-wavelength anomalous dispersion methods to solve the complex of the DNA-binding domain (DBD) of interferon regulatory factor 4 (IRF4) bound to its interferon-stimulated response element (ISRE). The structure unexpectedly shows three molecules of the IRF4 DBD bound to one ISRE. The sole reliance on native intrinsic anomalous scattering elements that belong to DNA-protein complexes renders the method of general applicability to a large number of such protein complexes that cannot be solved by molecular replacement or by other phasing methods.