Summary information and primary citation
- PDB-id
- 7oar; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase
- Method
- X-ray (2.58 Å)
- Summary
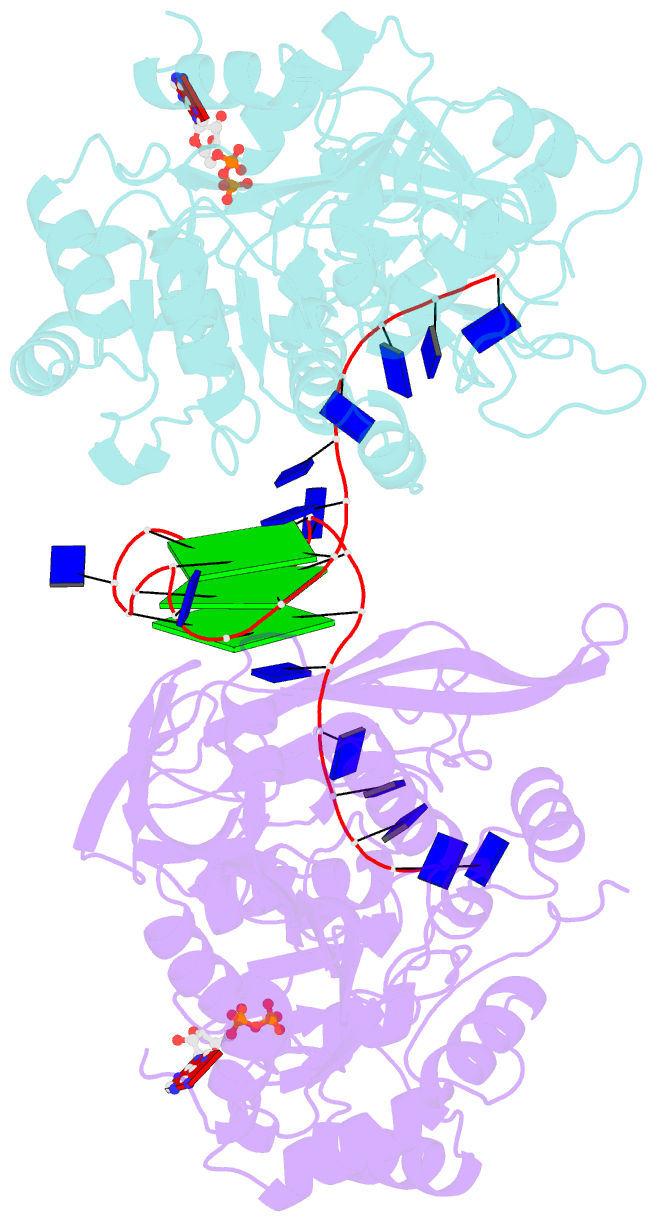
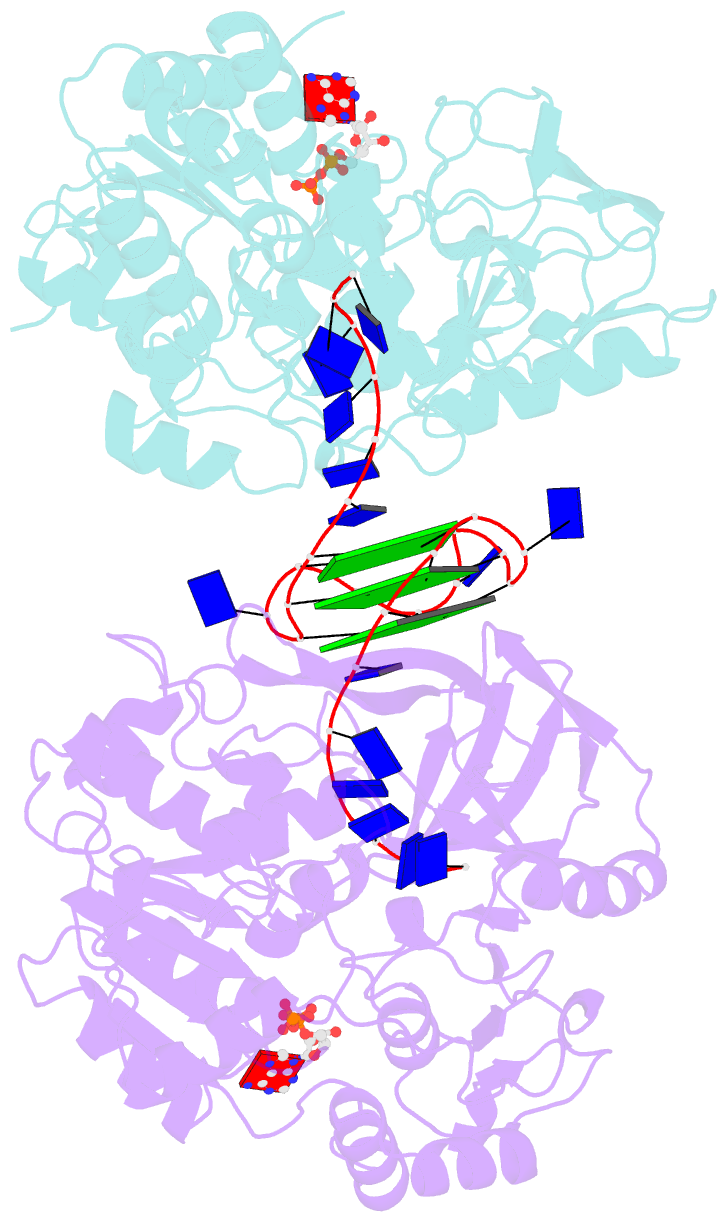
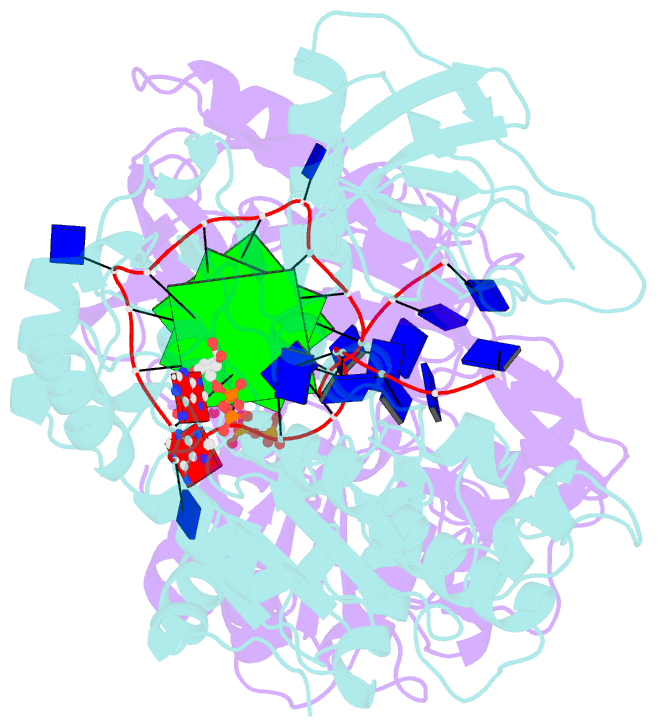
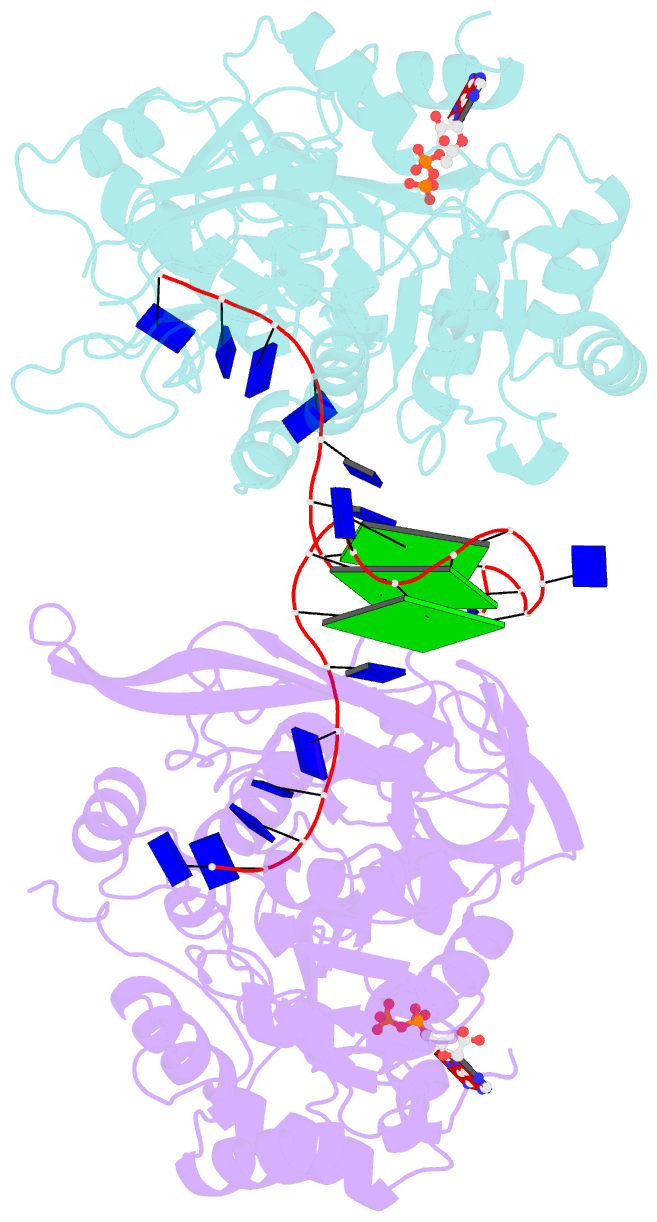
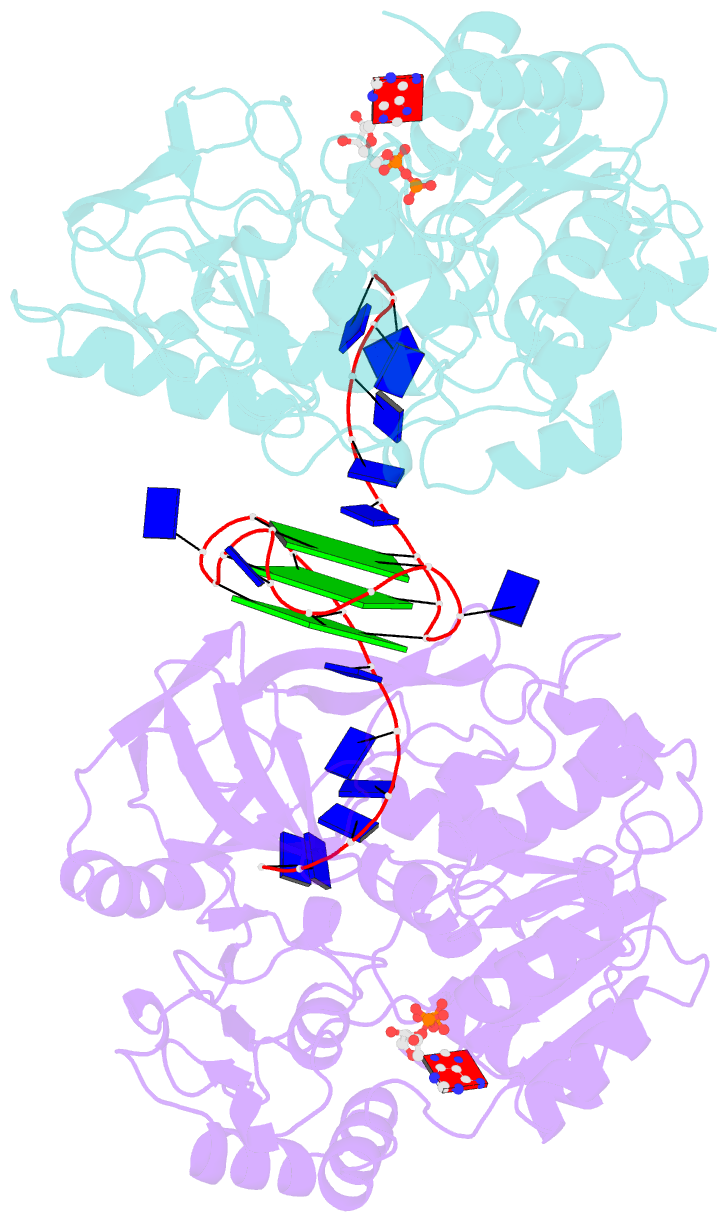
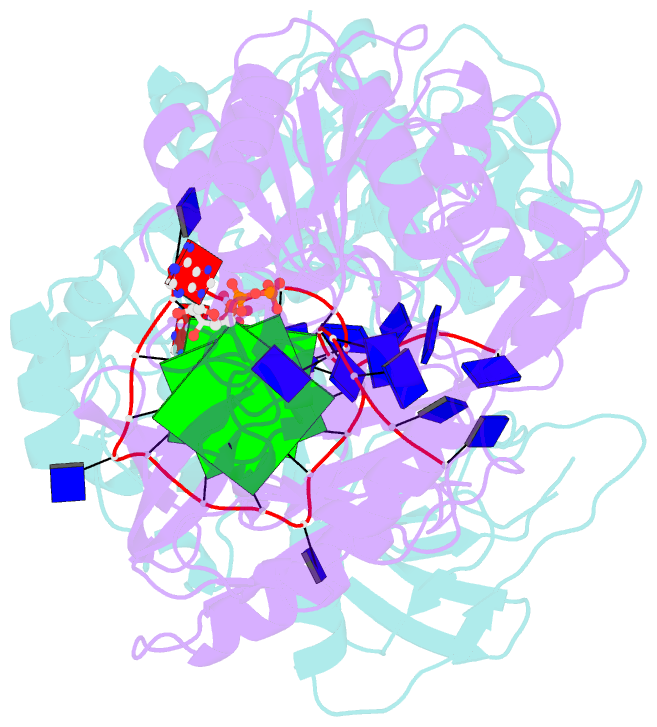
- Crystal structure of helicase pif1 from thermus oshimai in complex with parallel g-quadruplex
- Reference
- Dai YX, Guo HL, Liu NN, Chen WF, Ai X, Li HH, Sun B, Hou XM, Rety S, Xi XG (2022): "Structural mechanism underpinning Thermus oshimai Pif1-mediated G-quadruplex unfolding." Embo Rep., 23, e53874. doi: 10.15252/embr.202153874.
- Abstract
- G-quadruplexes (G4s) are unusual stable DNA structures that cause genomic instability. To overcome the potential barriers formed by G4s, cells have evolved different families of proteins that unfold G4s. Pif1 is a DNA helicase from superfamily 1 (SF1) conserved from bacteria to humans with high G4-unwinding activity. Here, we present the first X-ray crystal structure of the Thermus oshimai Pif1 (ToPif1) complexed with a G4. Our structure reveals that ToPif1 recognizes the entire native G4 via a cluster of amino acids at domains 1B/2B which constitute a G4-Recognizing Surface (GRS). The overall structure of the G4 maintains its three-layered propeller-type G4 topology, without significant reorganization of G-tetrads upon protein binding. The three G-tetrads in G4 are recognized by GRS residues mainly through electrostatic, ionic interactions, and hydrogen bonds formed between the GRS residues and the ribose-phosphate backbone. Compared with previously solved structures of SF2 helicases in complex with G4, our structure reveals how helicases from distinct superfamilies adopt different strategies for recognizing and unfolding G4s.