Summary information and primary citation
- PDB-id
- 7odf; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein
- Method
- cryo-EM (2.66 Å)
- Summary
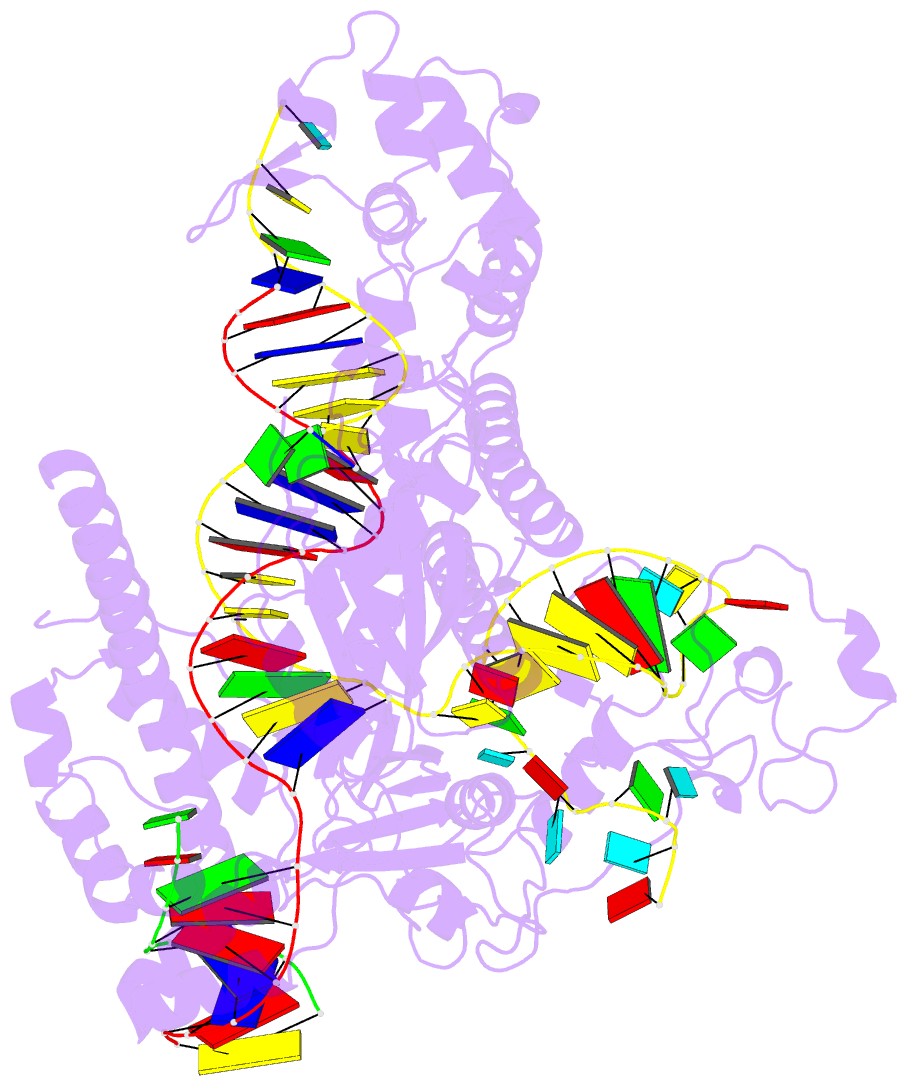
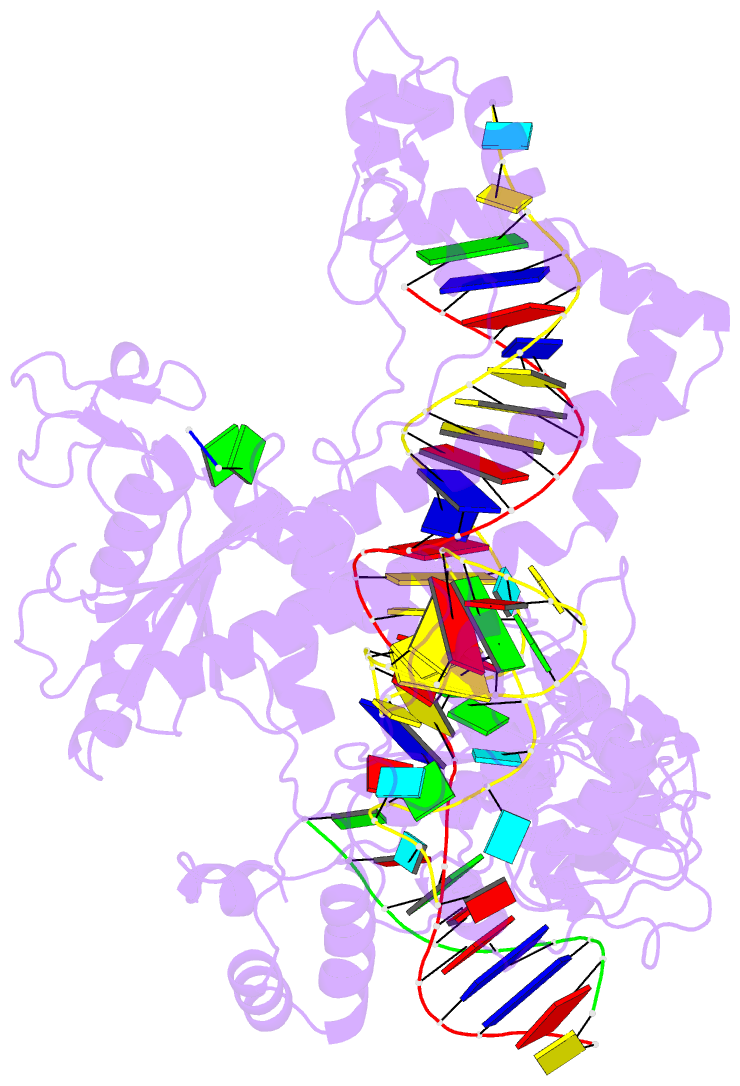
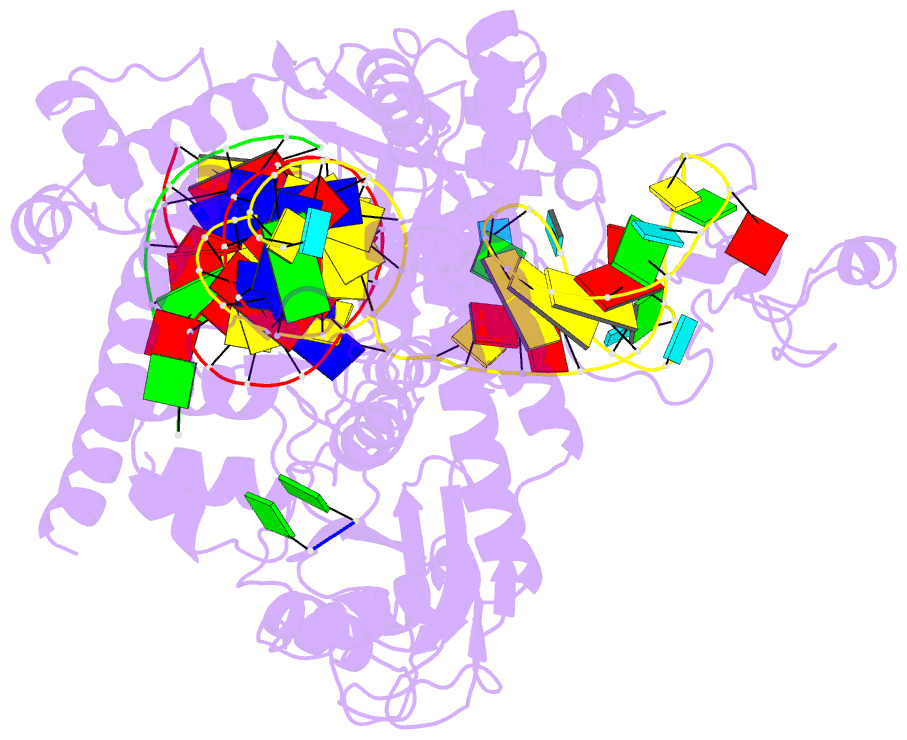
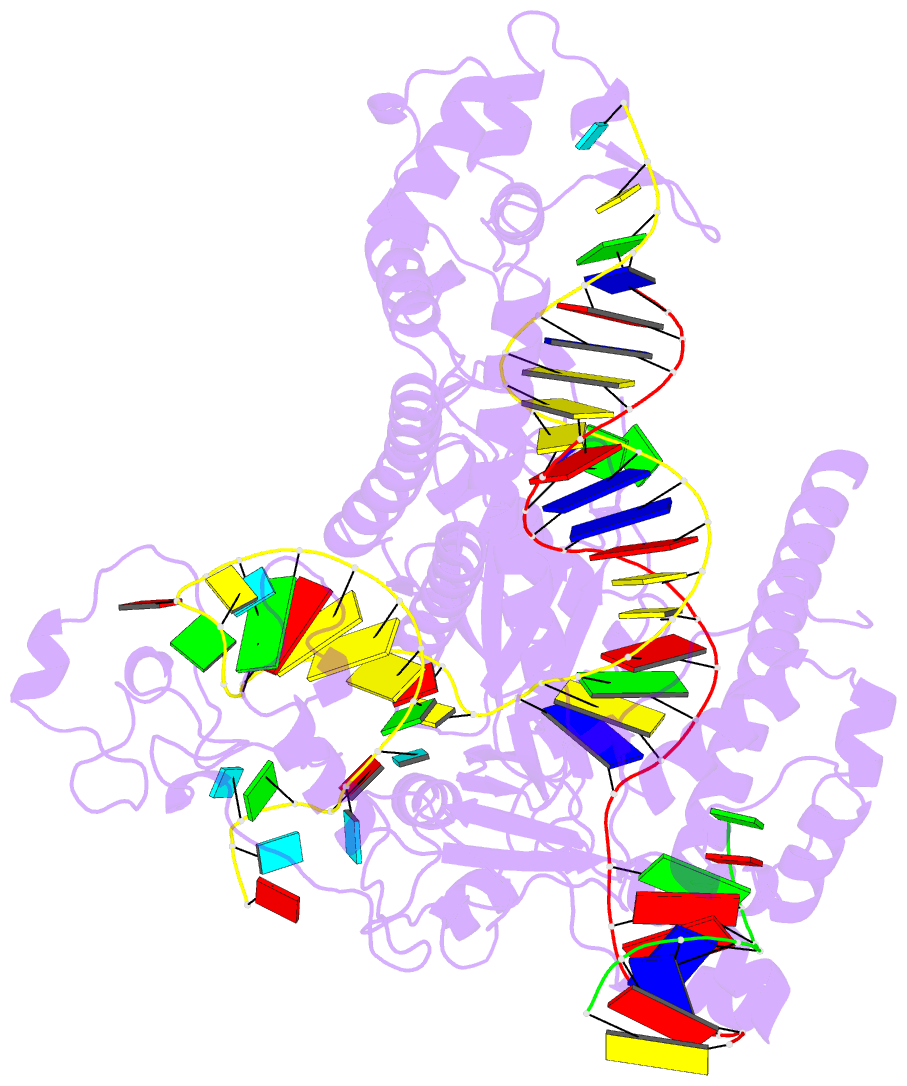
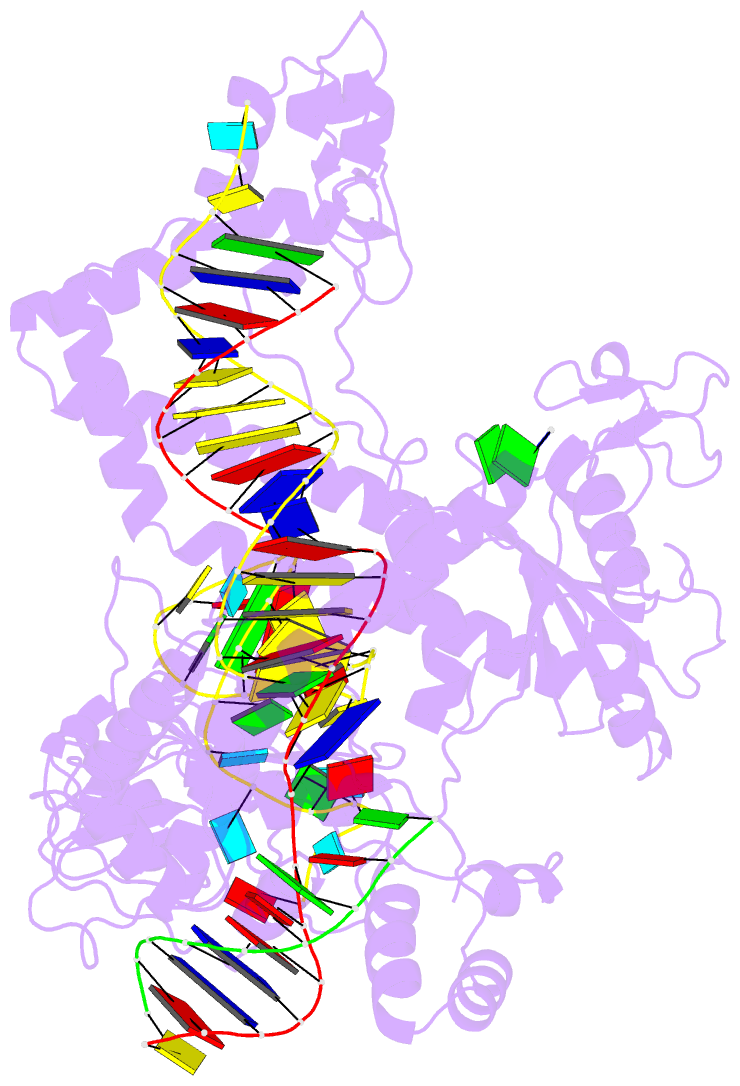
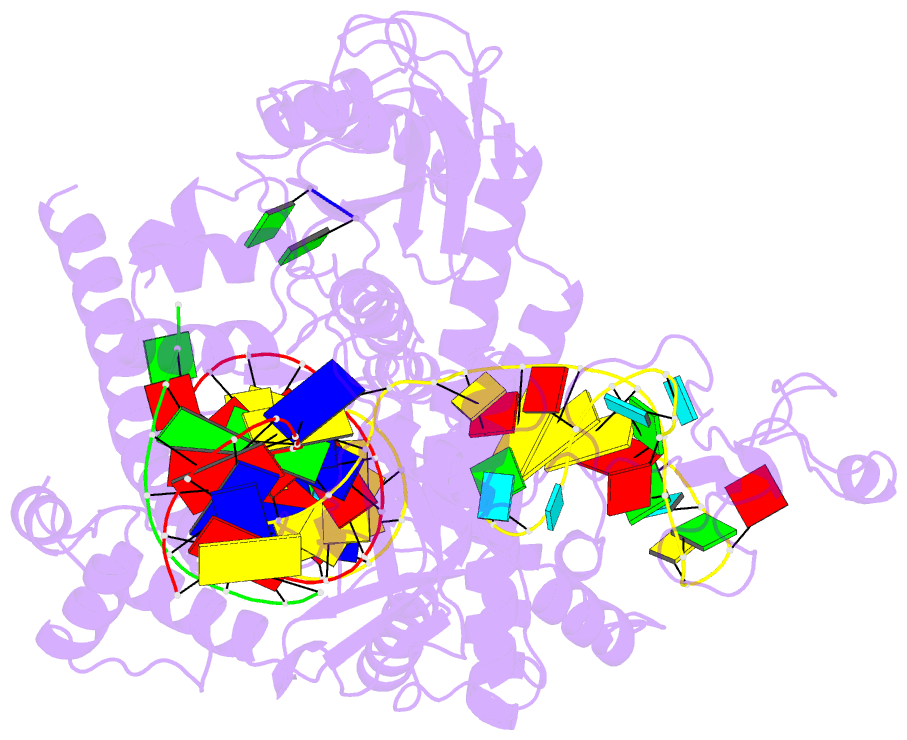
- Structure of the mini-RNA-guided endonuclease crispr-cas_phi3
- Reference
- Carabias A, Fuglsang A, Temperini P, Pape T, Sofos N, Stella S, Erlendsson S, Montoya G (2021): "Structure of the mini-RNA-guided endonuclease CRISPR-Cas12j3." Nat Commun, 12, 4476. doi: 10.1038/s41467-021-24707-3.
- Abstract
- CRISPR-Cas12j is a recently identified family of miniaturized RNA-guided endonucleases from phages. These ribonucleoproteins provide a compact scaffold gathering all key activities of a genome editing tool. We provide the first structural insight into the Cas12j family by determining the cryoEM structure of Cas12j3/R-loop complex after DNA cleavage. The structure reveals the machinery for PAM recognition, hybrid assembly and DNA cleavage. The crRNA-DNA hybrid is directed to the stop domain that splits the hybrid, guiding the T-strand towards the catalytic site. The conserved RuvC insertion is anchored in the stop domain and interacts along the phosphate backbone of the crRNA in the hybrid. The assembly of a hybrid longer than 12-nt activates catalysis through key functional residues in the RuvC insertion. Our findings suggest why Cas12j unleashes unspecific ssDNA degradation after activation. A site-directed mutagenesis analysis supports the DNA cutting mechanism, providing new avenues to redesign CRISPR-Cas12j nucleases for genome editing.