Summary information and primary citation
- PDB-id
- 7ol9; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- X-ray (2.9 Å)
- Summary
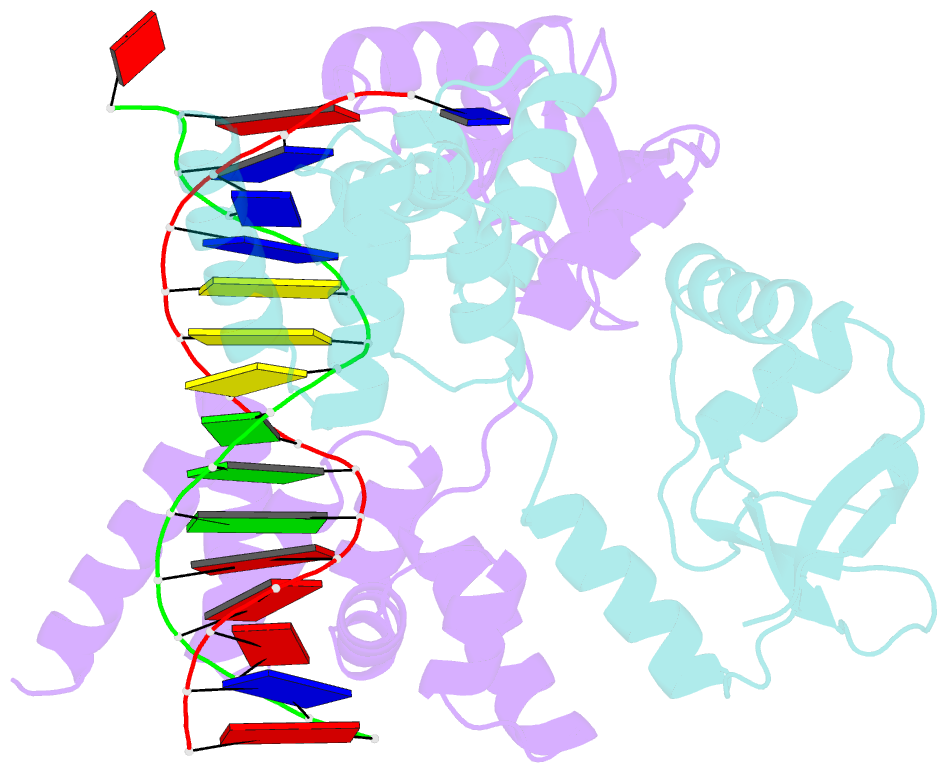
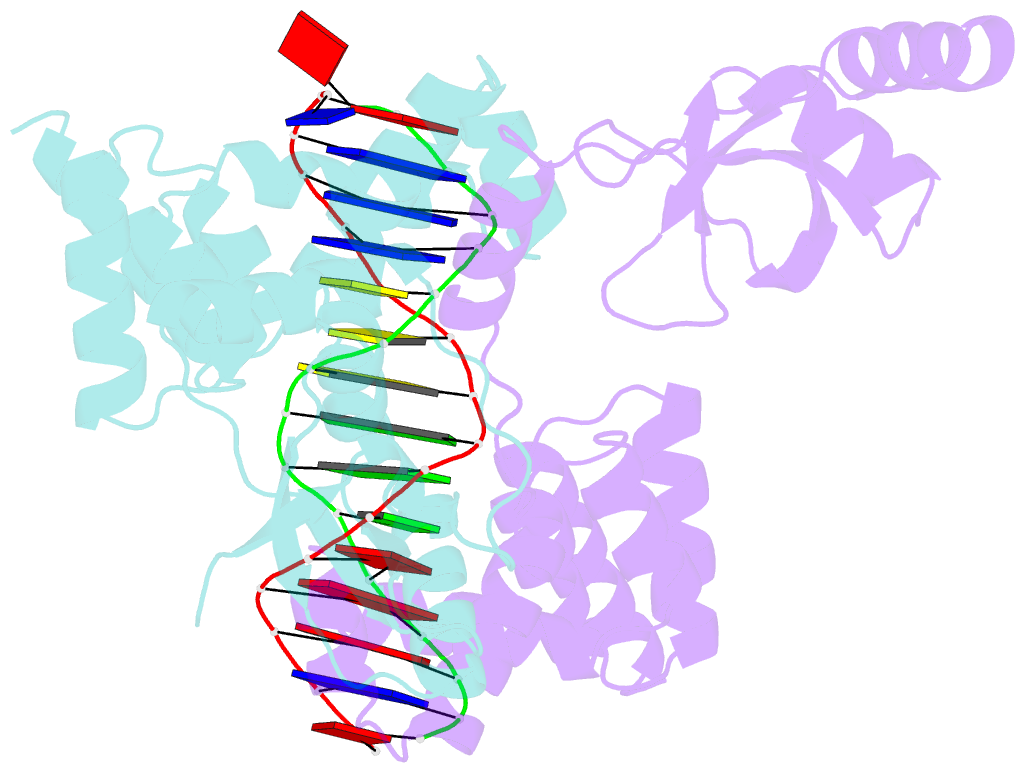
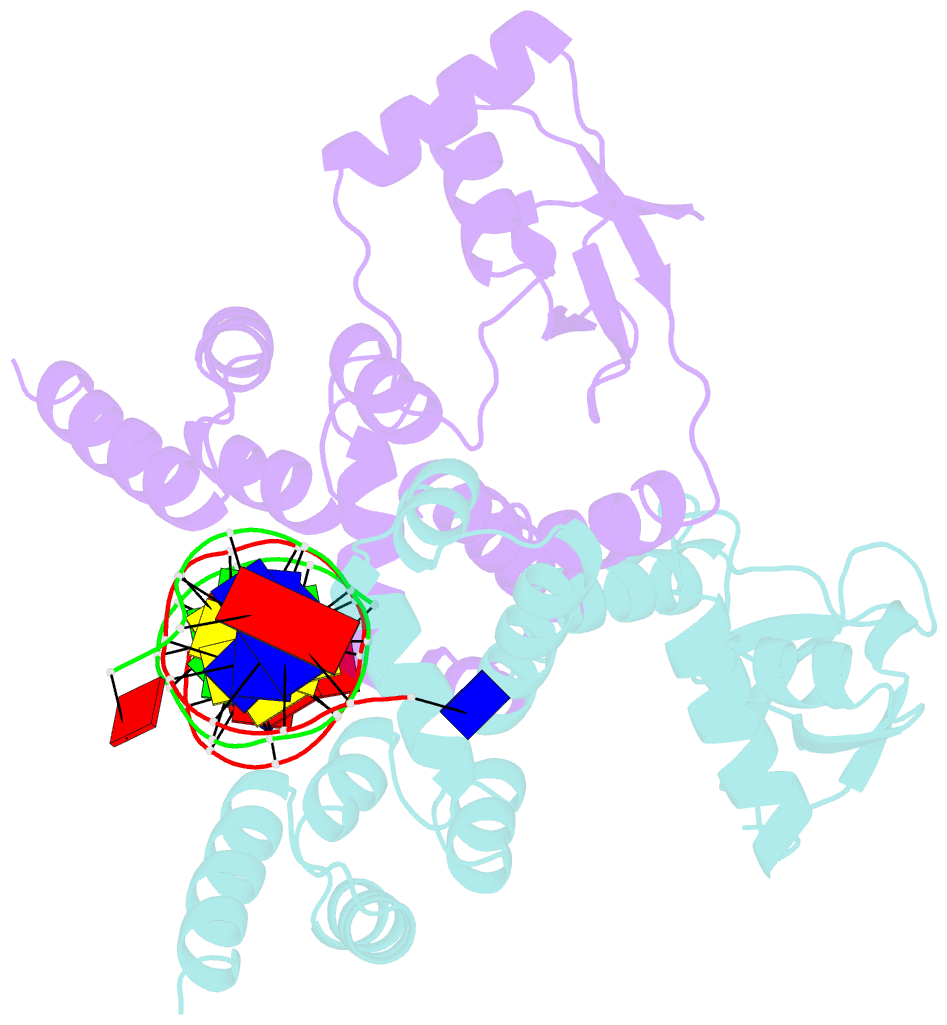
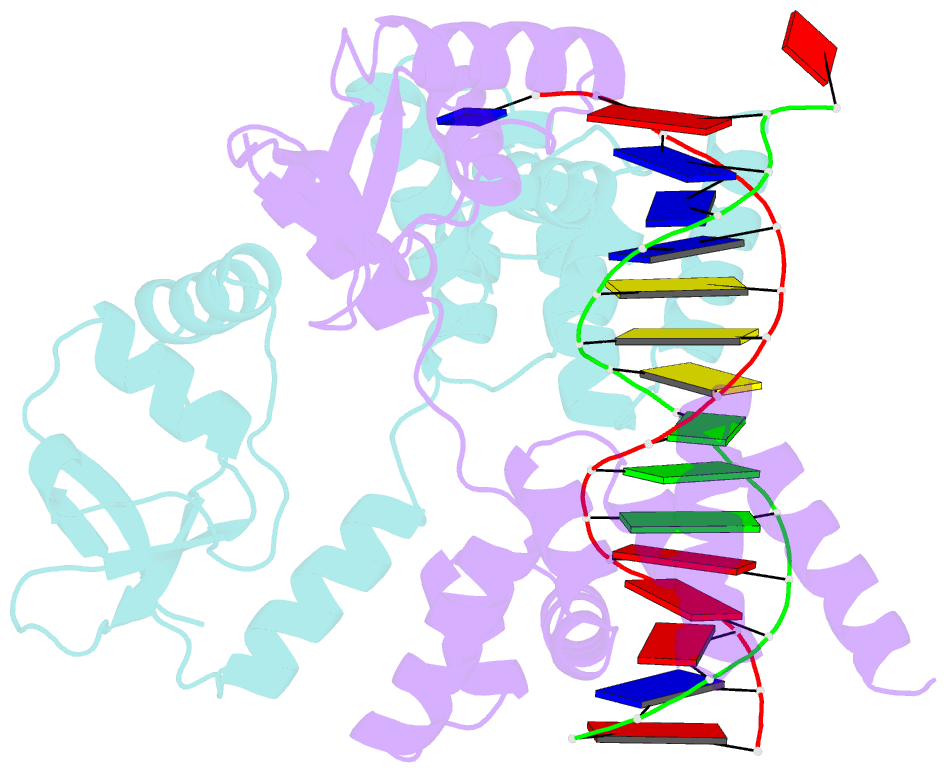
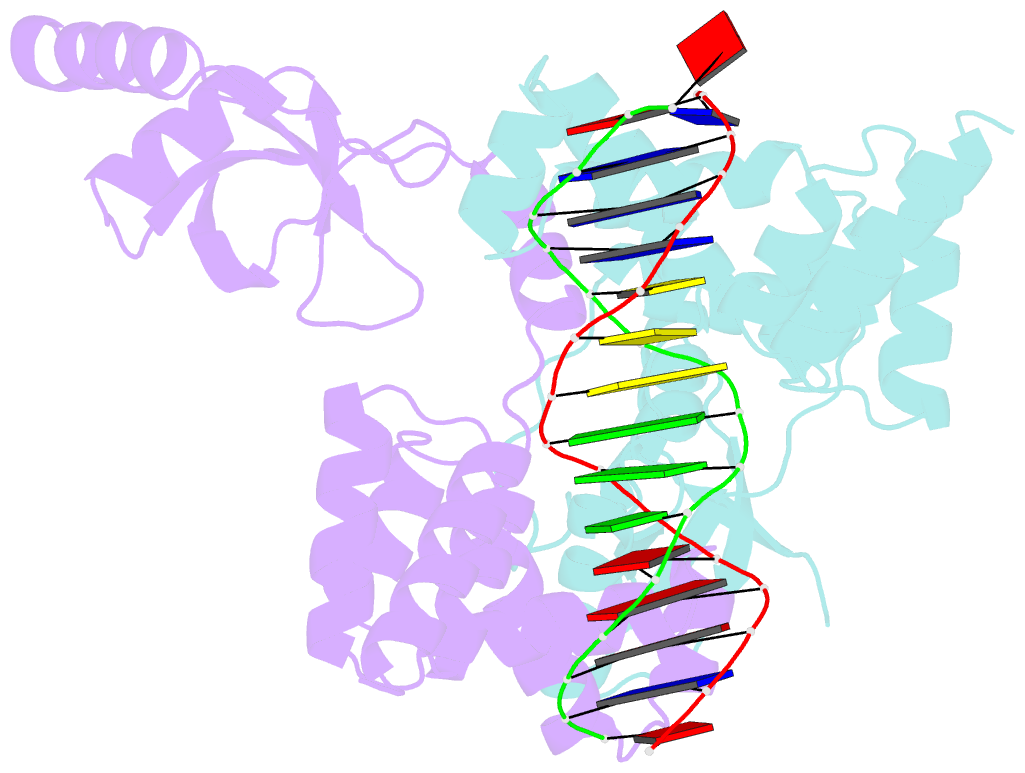
- Crystal structure of c-terminally truncated bacillus subtilis nucleoid occlusion protein (noc) complexed to the noc-binding site (nbs)
- Reference
- Sukhoverkov KV, Jalal ASB, Ault JR, Sobott F, Lawson DM, Le TBK (2023): "The CTP-binding domain is disengaged from the DNA-binding domain in a cocrystal structure of Bacillus subtilis Noc-DNA complex." J.Biol.Chem., 299, 103063. doi: 10.1016/j.jbc.2023.103063.
- Abstract
- In Bacillus subtilis, a ParB-like nucleoid occlusion protein (Noc) binds specifically to Noc-binding sites (NBS) on the chromosome to help coordinate chromosome segregation and cell division. Noc does so by binding to cytidine triphosphate (CTP) to form large membrane-associated nucleoprotein complexes to physically inhibit the assembly of the cell division machinery. The site-specific binding of Noc to NBS DNA is a prerequisite for CTP-binding and the subsequent formation of a membrane-active DNA-entrapped protein complex. Here, we solve the structure of a C-terminally truncated B. subtilis Noc bound to NBS DNA to reveal the conformation of Noc at this crucial step. Our structure reveals the disengagement between the N-terminal CTP-binding domain and the NBS-binding domain of each DNA-bound Noc subunit, this is driven, in part, by the swapping of helices 4 and 5 at the interface of the two domains. Site-specific crosslinking data suggest that this conformation of Noc-NBS exists in solution. Overall, our results lend support to the recent proposal that parS/NBS-binding catalyzes CTP-binding and DNA-entrapment by preventing the re-engagement of the CTP-binding domain and the DNA-binding domain from the same ParB/Noc subunit.