Summary information and primary citation
- PDB-id
- 7onu; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein
- Method
- cryo-EM (3.0 Å)
- Summary
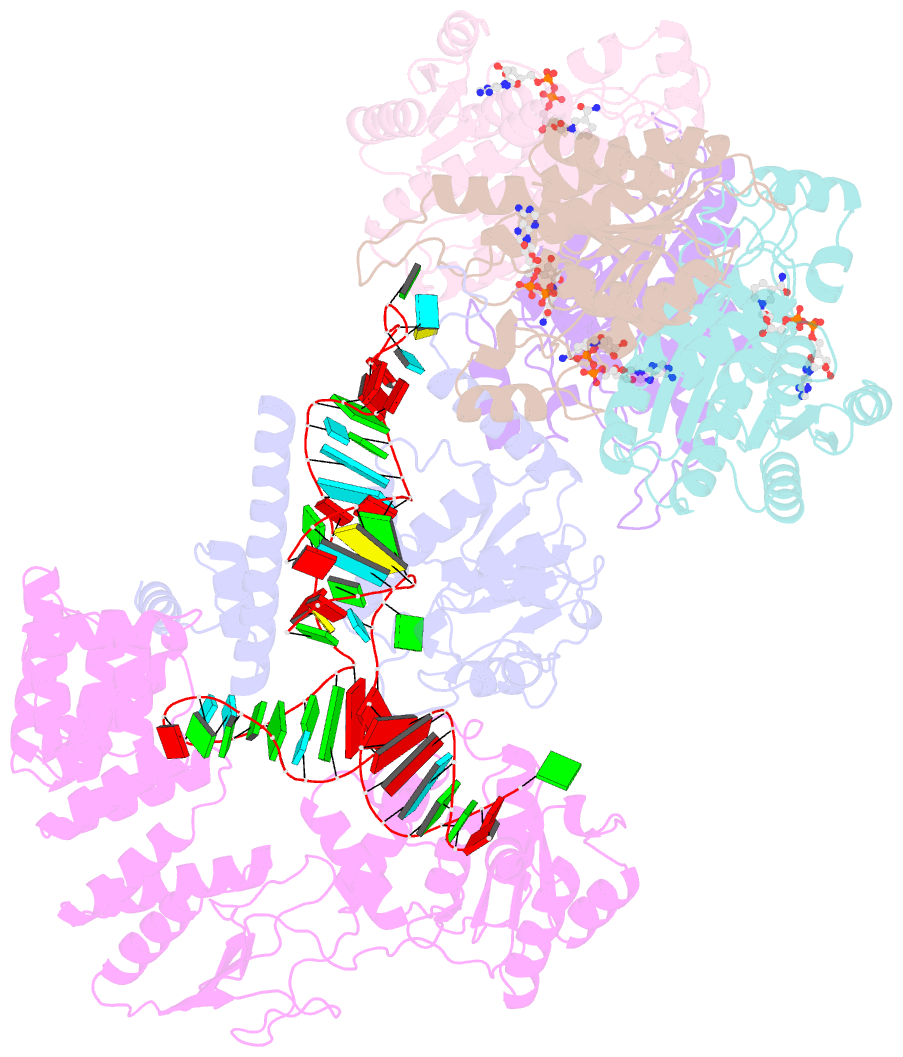
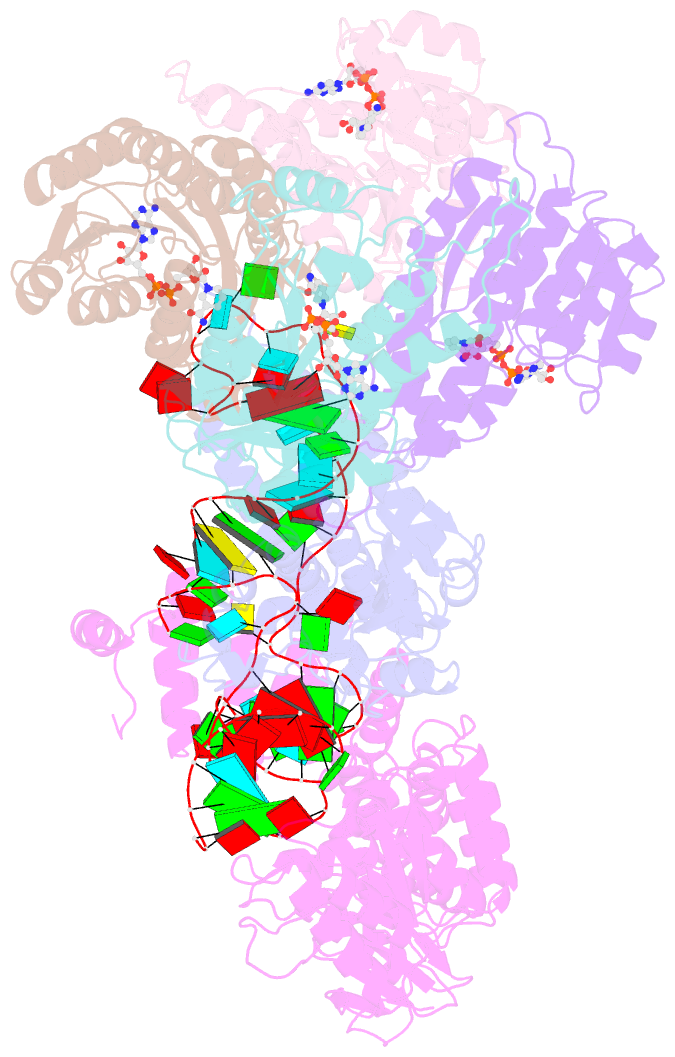
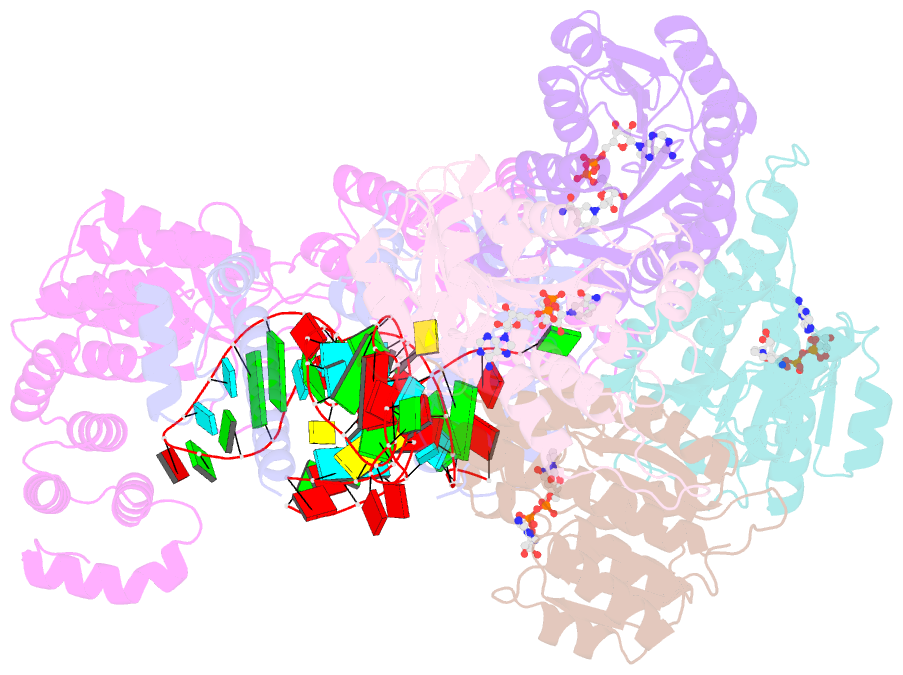
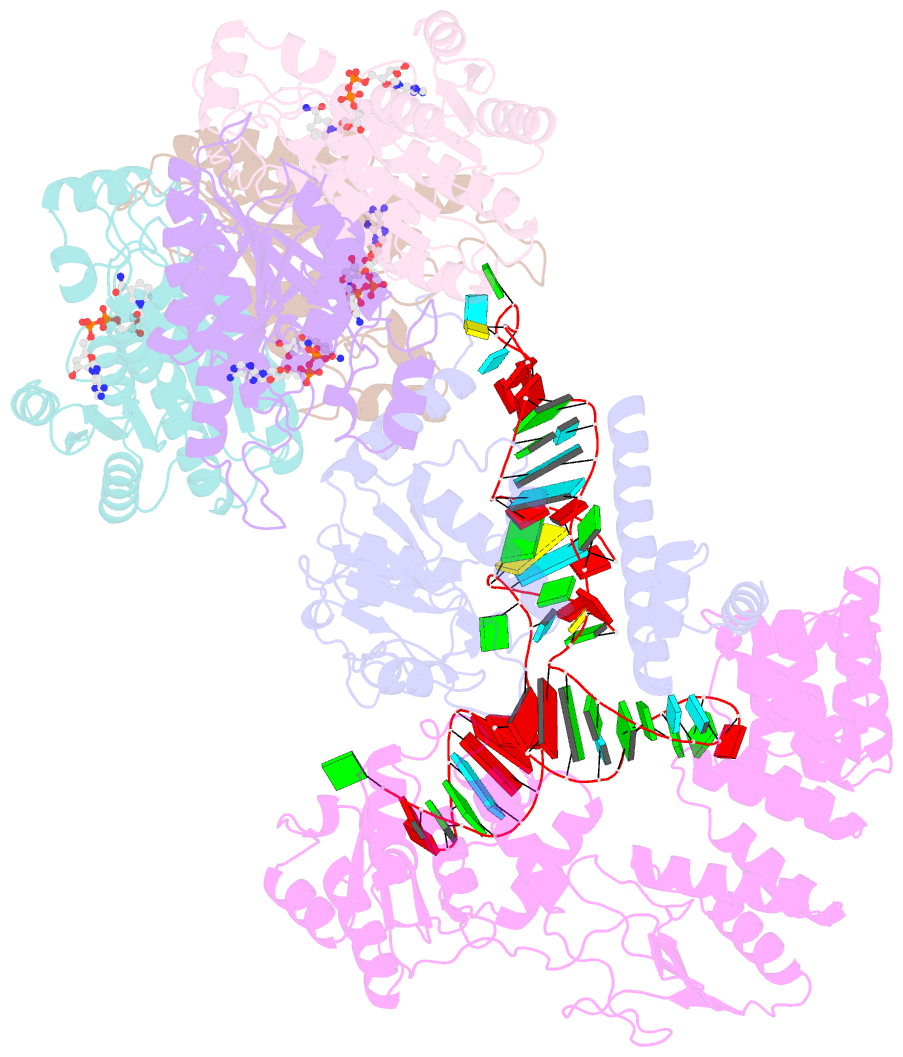
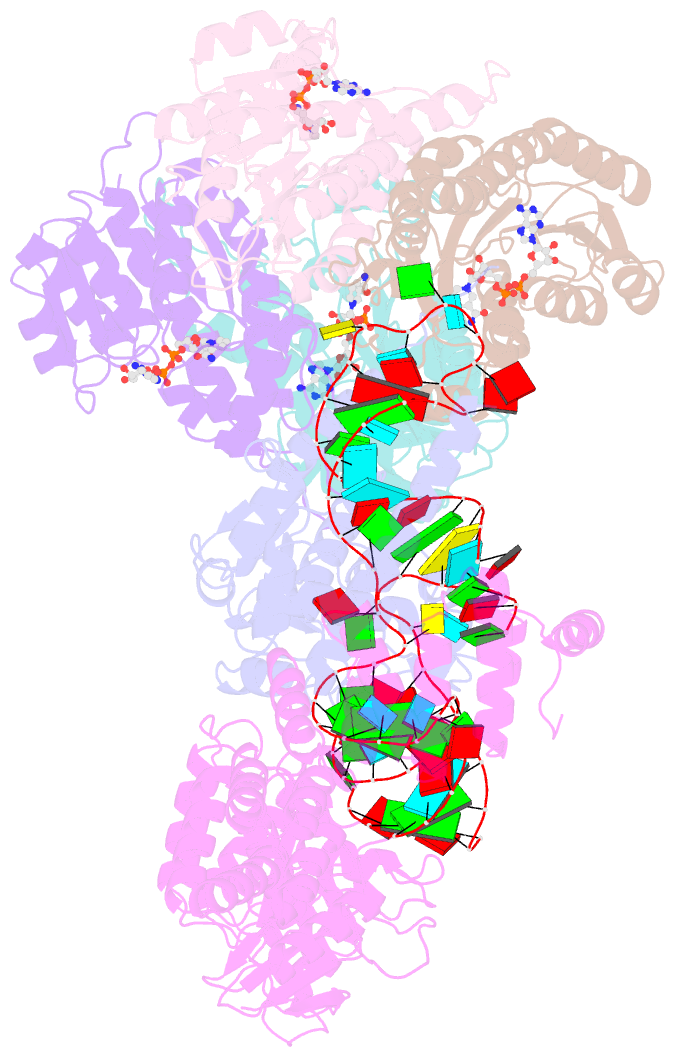
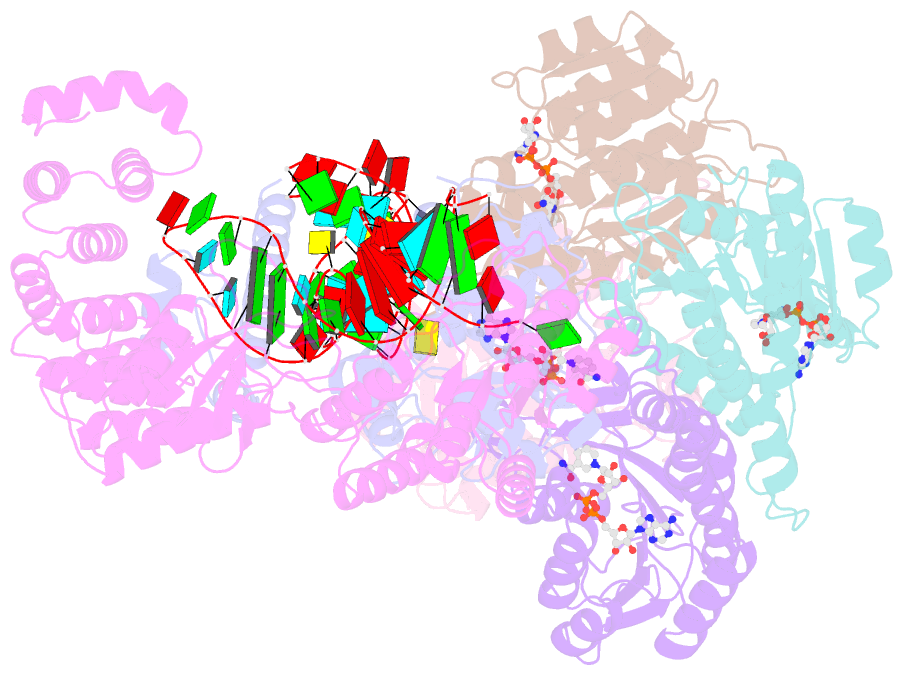
- Structure of human mitochondrial rnase p in complex with mitochondrial pre-trna-tyr
- Reference
- Bhatta A, Dienemann C, Cramer P, Hillen HS (2021): "Structural basis of RNA processing by human mitochondrial RNase P." Nat.Struct.Mol.Biol., 28, 713-723. doi: 10.1038/s41594-021-00637-y.
- Abstract
- Human mitochondrial transcripts contain messenger and ribosomal RNAs flanked by transfer RNAs (tRNAs), which are excised by mitochondrial RNase (mtRNase) P and Z to liberate all RNA species. In contrast to nuclear or bacterial RNase P, mtRNase P is not a ribozyme but comprises three protein subunits that carry out RNA cleavage and methylation by unknown mechanisms. Here, we present the cryo-EM structure of human mtRNase P bound to precursor tRNA, which reveals a unique mechanism of substrate recognition and processing. Subunits TRMT10C and SDR5C1 form a subcomplex that binds conserved mitochondrial tRNA elements, including the anticodon loop, and positions the tRNA for methylation. The endonuclease PRORP is recruited and activated through interactions with its PPR and nuclease domains to ensure precise pre-tRNA cleavage. The structure provides the molecular basis for the first step of RNA processing in human mitochondria.