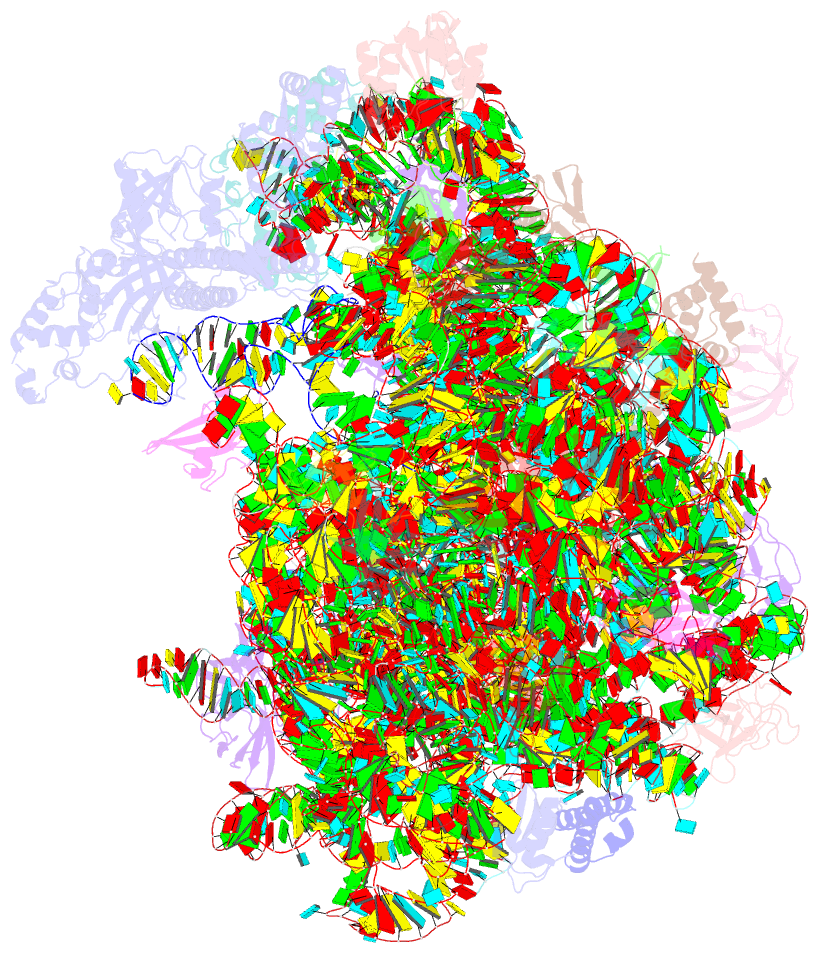
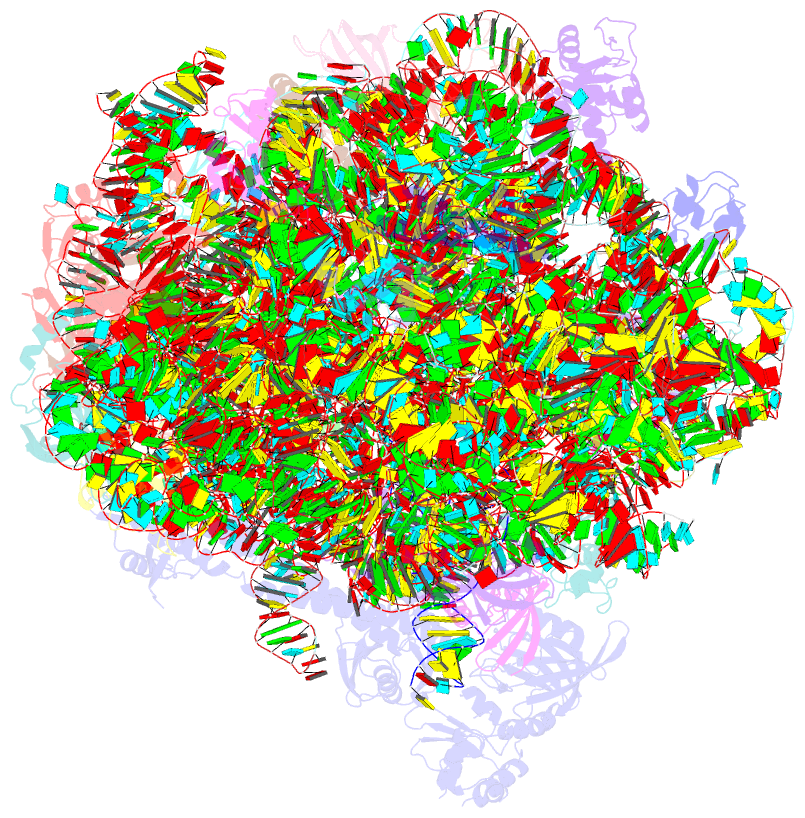
Summary information and primary citation
- PDB-id
- 7ope; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ribosome
- Method
- cryo-EM (3.2 Å)
- Summary
- Rqch dr variant bound to 50s-peptidyl-trna-rqcp rqc complex (rigid body refinement)
- Reference
- Takada H, Crowe-McAuliffe C, Polte C, Sidorova ZY, Murina V, Atkinson GC, Konevega AL, Ignatova Z, Wilson DN, Hauryliuk V (2021): "RqcH and RqcP catalyze processive poly-alanine synthesis in a reconstituted ribosome-associated quality control system." Nucleic Acids Res., 49, 8355-8369. doi: 10.1093/nar/gkab589.
- Abstract
- In the cell, stalled ribosomes are rescued through ribosome-associated protein quality-control (RQC) pathways. After splitting of the stalled ribosome, a C-terminal polyalanine 'tail' is added to the unfinished polypeptide attached to the tRNA on the 50S ribosomal subunit. In Bacillus subtilis, polyalanine tailing is catalyzed by the NEMF family protein RqcH, in cooperation with RqcP. However, the mechanistic details of this process remain unclear. Here we demonstrate that RqcH is responsible for tRNAAla selection during RQC elongation, whereas RqcP lacks any tRNA specificity. The ribosomal protein uL11 is crucial for RqcH, but not RqcP, recruitment to the 50S subunit, and B. subtilis lacking uL11 are RQC-deficient. Through mutational mapping, we identify critical residues within RqcH and RqcP that are important for interaction with the P-site tRNA and/or the 50S subunit. Additionally, we have reconstituted polyalanine-tailing in vitro and can demonstrate that RqcH and RqcP are necessary and sufficient for processivity in a minimal system. Moreover, the in vitro reconstituted system recapitulates our in vivo findings by reproducing the importance of conserved residues of RqcH and RqcP for functionality. Collectively, our findings provide mechanistic insight into the role of RqcH and RqcP in the bacterial RQC pathway.