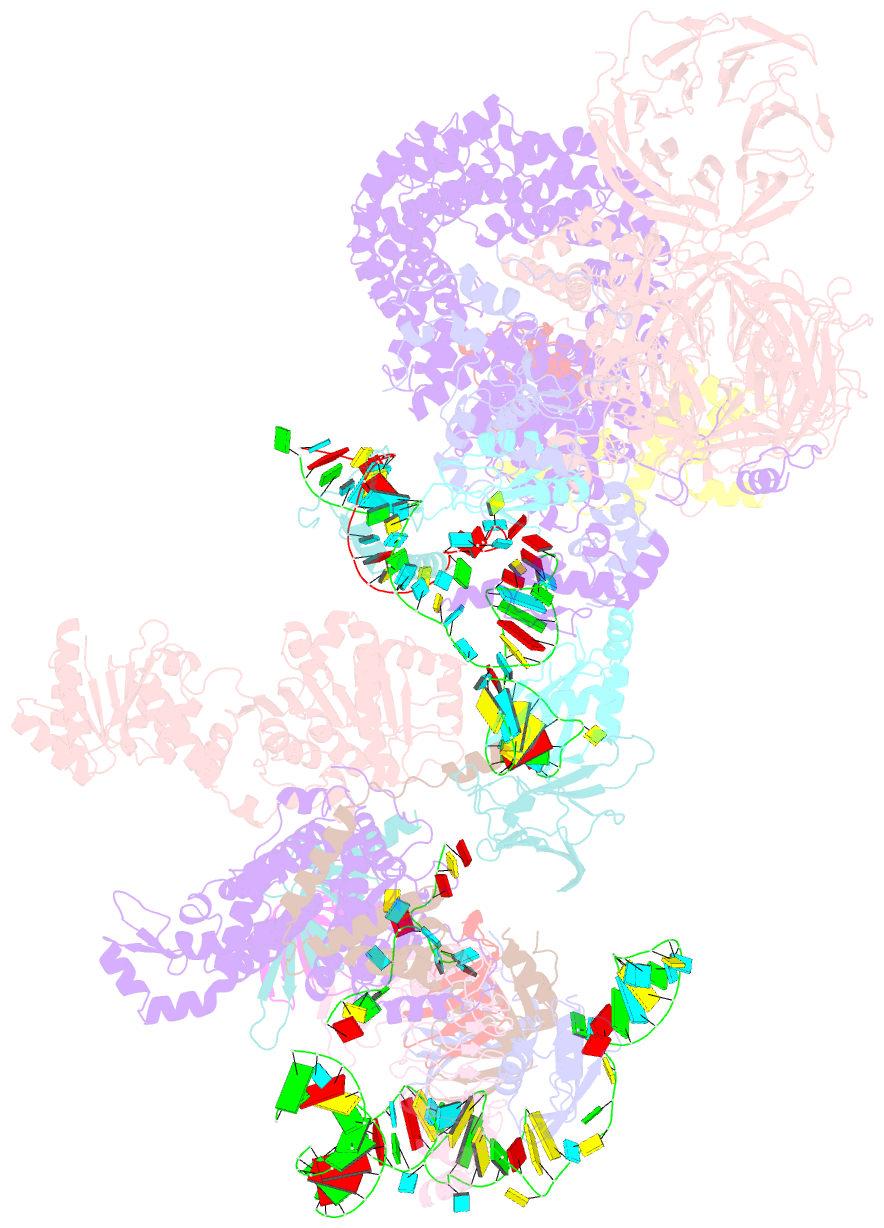
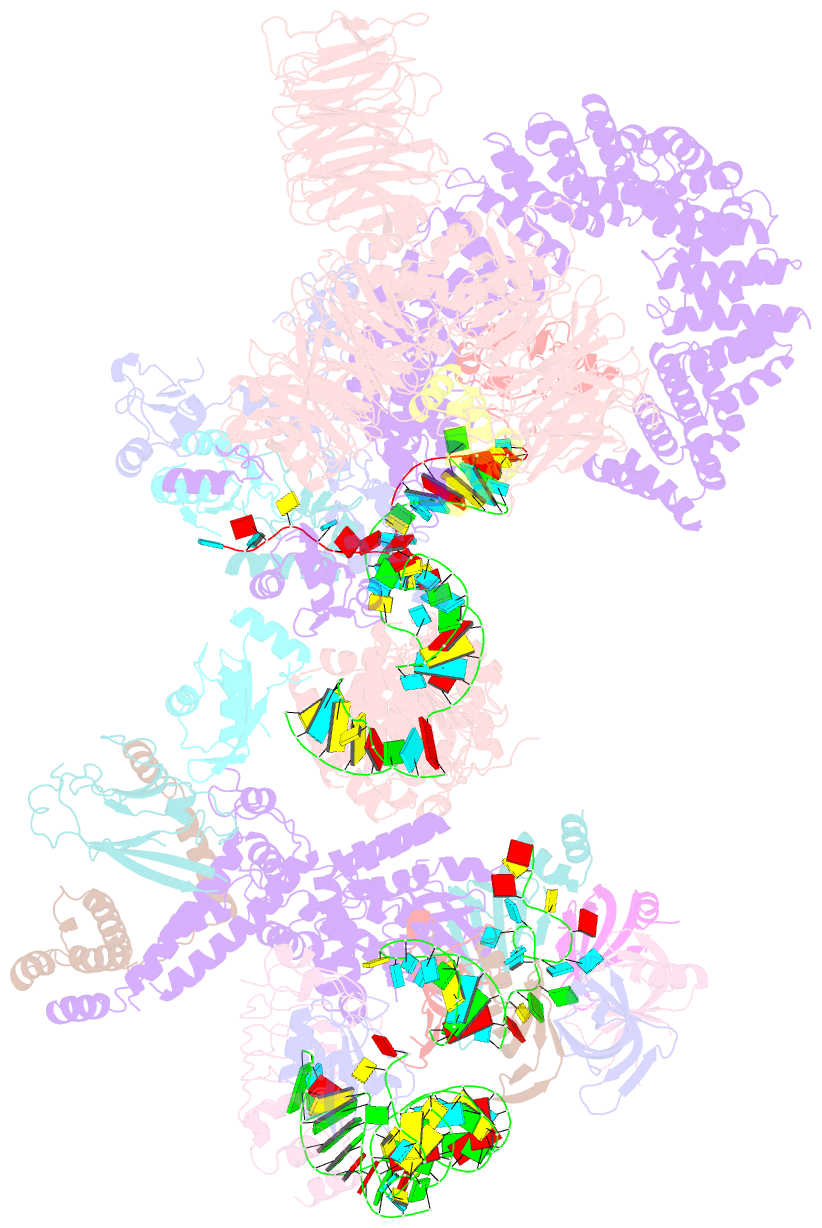
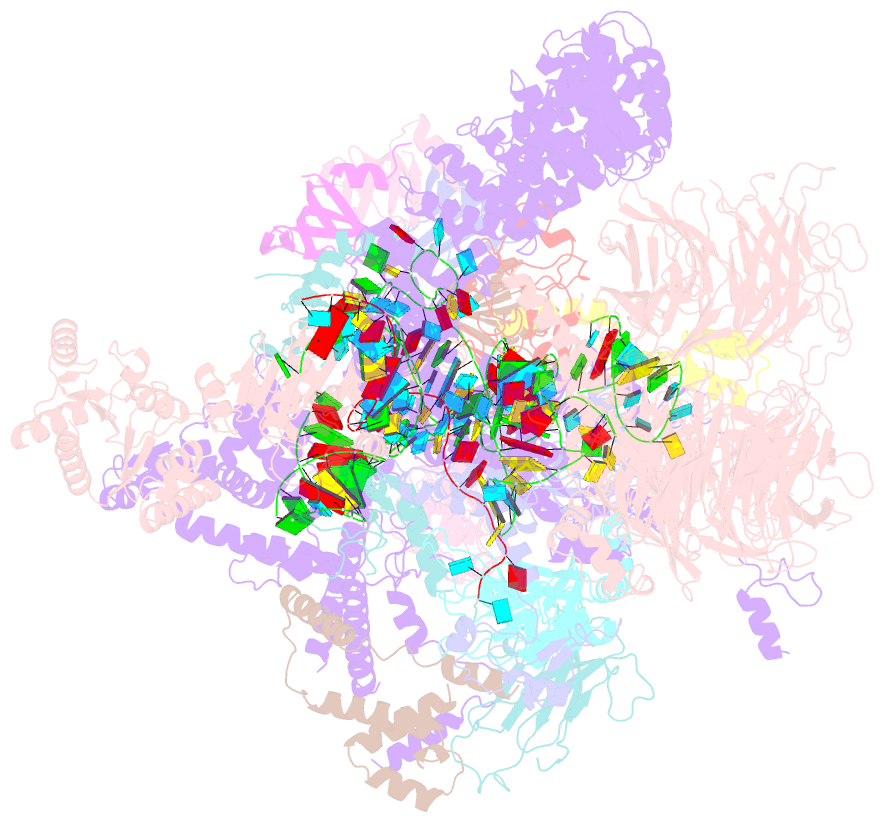
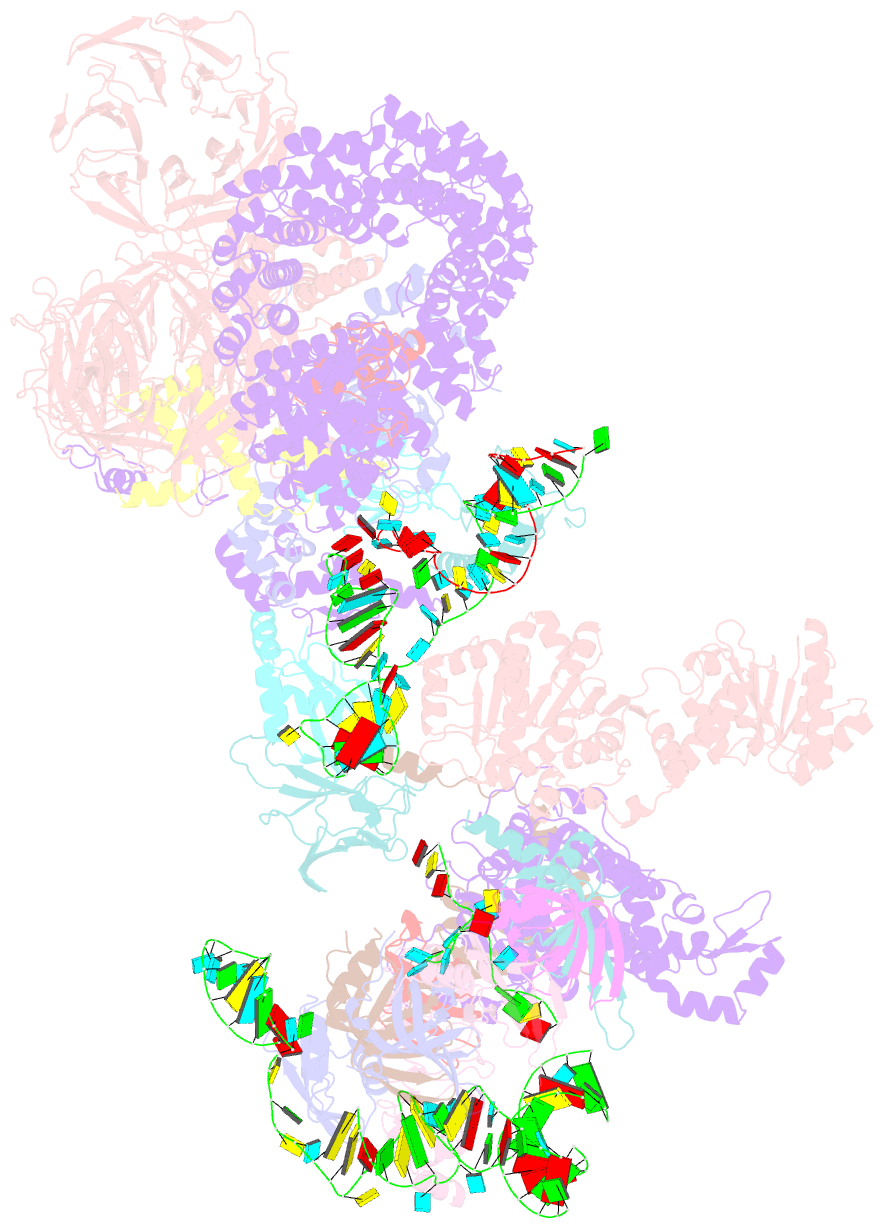
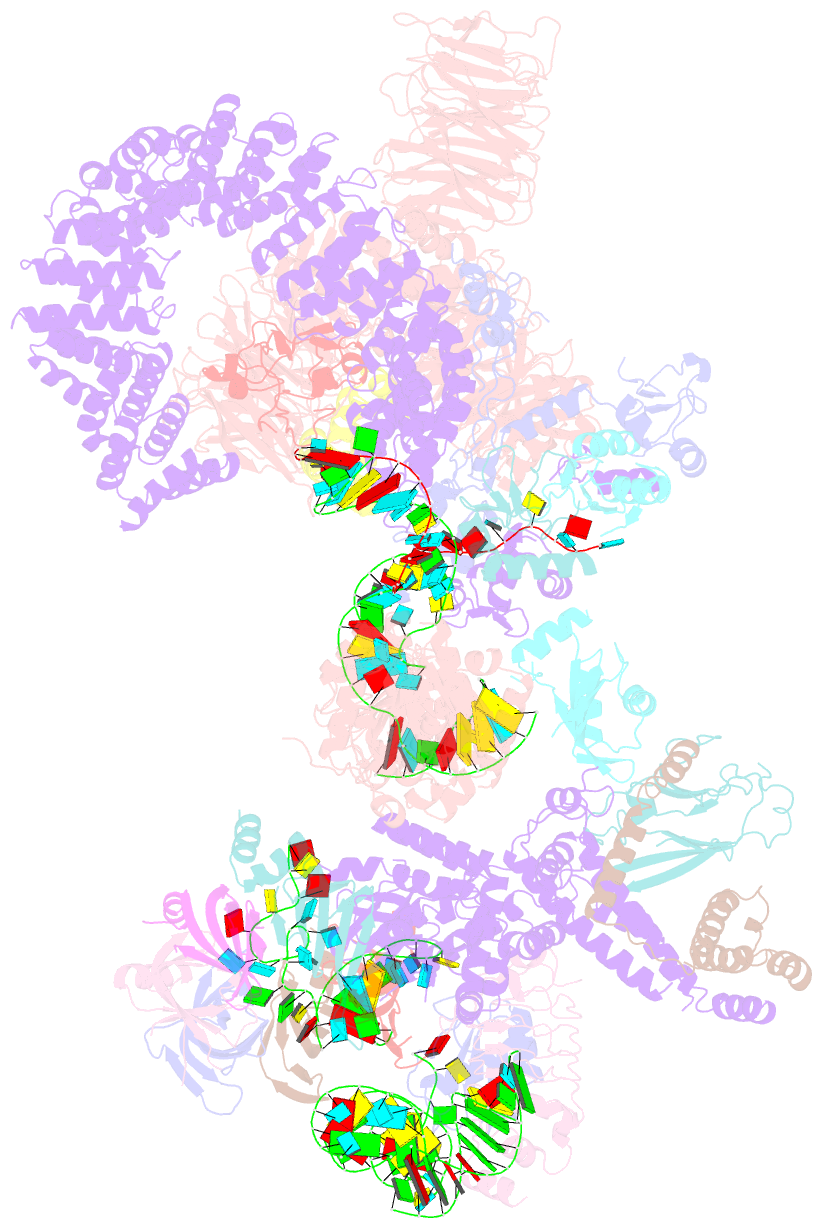
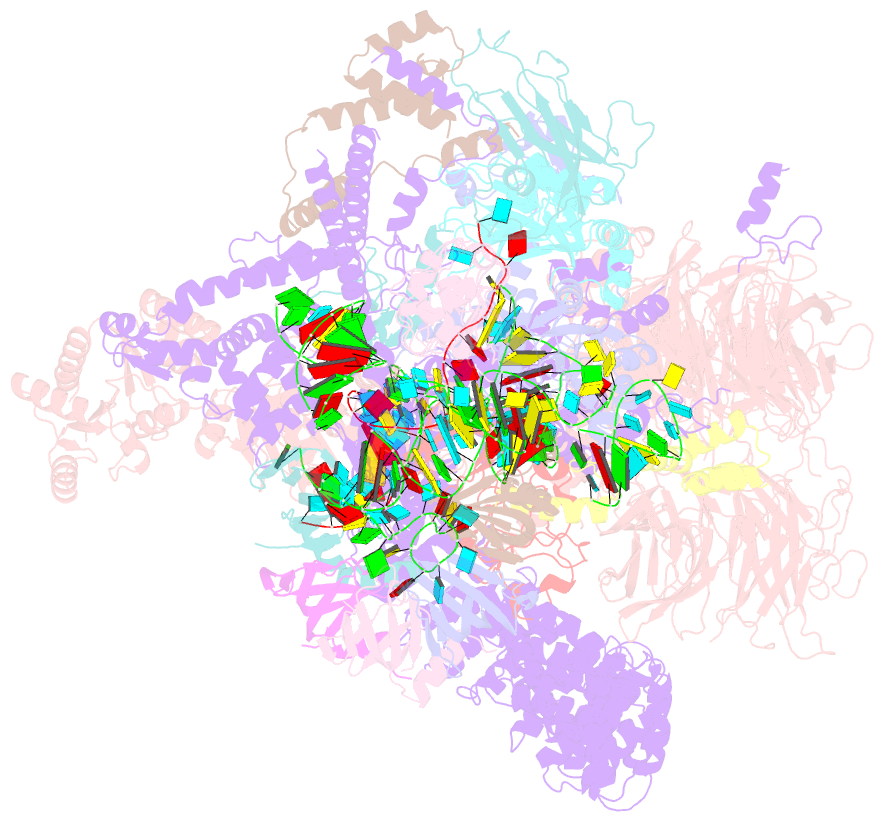
Summary information and primary citation
- PDB-id
- 7oqb; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- splicing
- Method
- cryo-EM (9.0 Å)
- Summary
- The u2 part of saccharomyces cerevisiae spliceosomal pre-a complex (delta bs-a act1)
- Reference
- Zhang Z, Rigo N, Dybkov O, Fourmann JB, Will CL, Kumar V, Urlaub H, Stark H, Luhrmann R (2021): "Structural insights into how Prp5 proofreads the pre-mRNA branch site." Nature, 596, 296-300. doi: 10.1038/s41586-021-03789-5.
- Abstract
- During the splicing of introns from precursor messenger RNAs (pre-mRNAs), the U2 small nuclear ribonucleoprotein (snRNP) must undergo stable integration into the spliceosomal A complex-a poorly understood, multistep process that is facilitated by the DEAD-box helicase Prp5 (refs. 1-4). During this process, the U2 small nuclear RNA (snRNA) forms an RNA duplex with the pre-mRNA branch site (the U2-BS helix), which is proofread by Prp5 at this stage through an unclear mechanism5. Here, by deleting the branch-site adenosine (BS-A) or mutating the branch-site sequence of an actin pre-mRNA, we stall the assembly of spliceosomes in extracts from the yeast Saccharomyces cerevisiae directly before the A complex is formed. We then determine the three-dimensional structure of this newly identified assembly intermediate by cryo-electron microscopy. Our structure indicates that the U2-BS helix has formed in this pre-A complex, but is not yet clamped by the HEAT domain of the Hsh155 protein (Hsh155HEAT), which exhibits an open conformation. The structure further reveals a large-scale remodelling/repositioning of the U1 and U2 snRNPs during the formation of the A complex that is required to allow subsequent binding of the U4/U6.U5 tri-snRNP, but that this repositioning is blocked in the pre-A complex by the presence of Prp5. Our data suggest that binding of Hsh155HEAT to the bulged BS-A of the U2-BS helix triggers closure of Hsh155HEAT, which in turn destabilizes Prp5 binding. Thus, Prp5 proofreads the branch site indirectly, hindering spliceosome assembly if branch-site mutations prevent the remodelling of Hsh155HEAT. Our data provide structural insights into how a spliceosomal helicase enhances the fidelity of pre-mRNA splicing.