Summary information and primary citation
- PDB-id
- 7orm; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein
- Method
- cryo-EM (3.3 Å)
- Summary
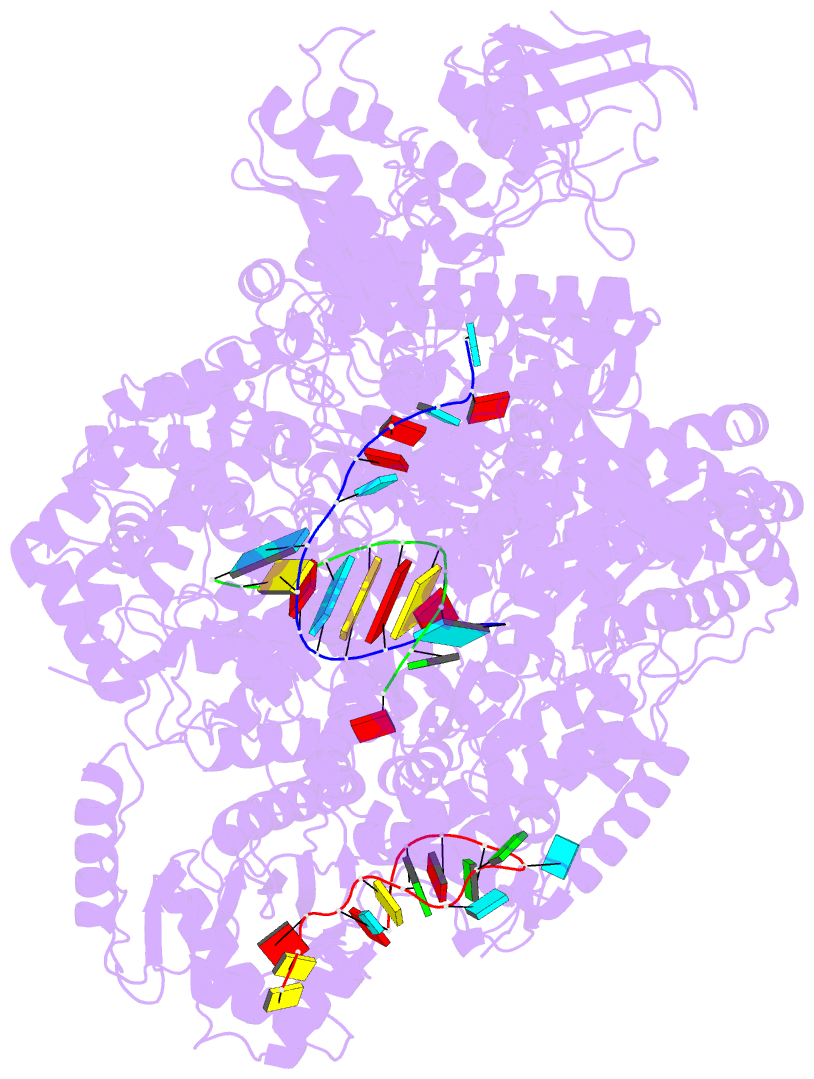
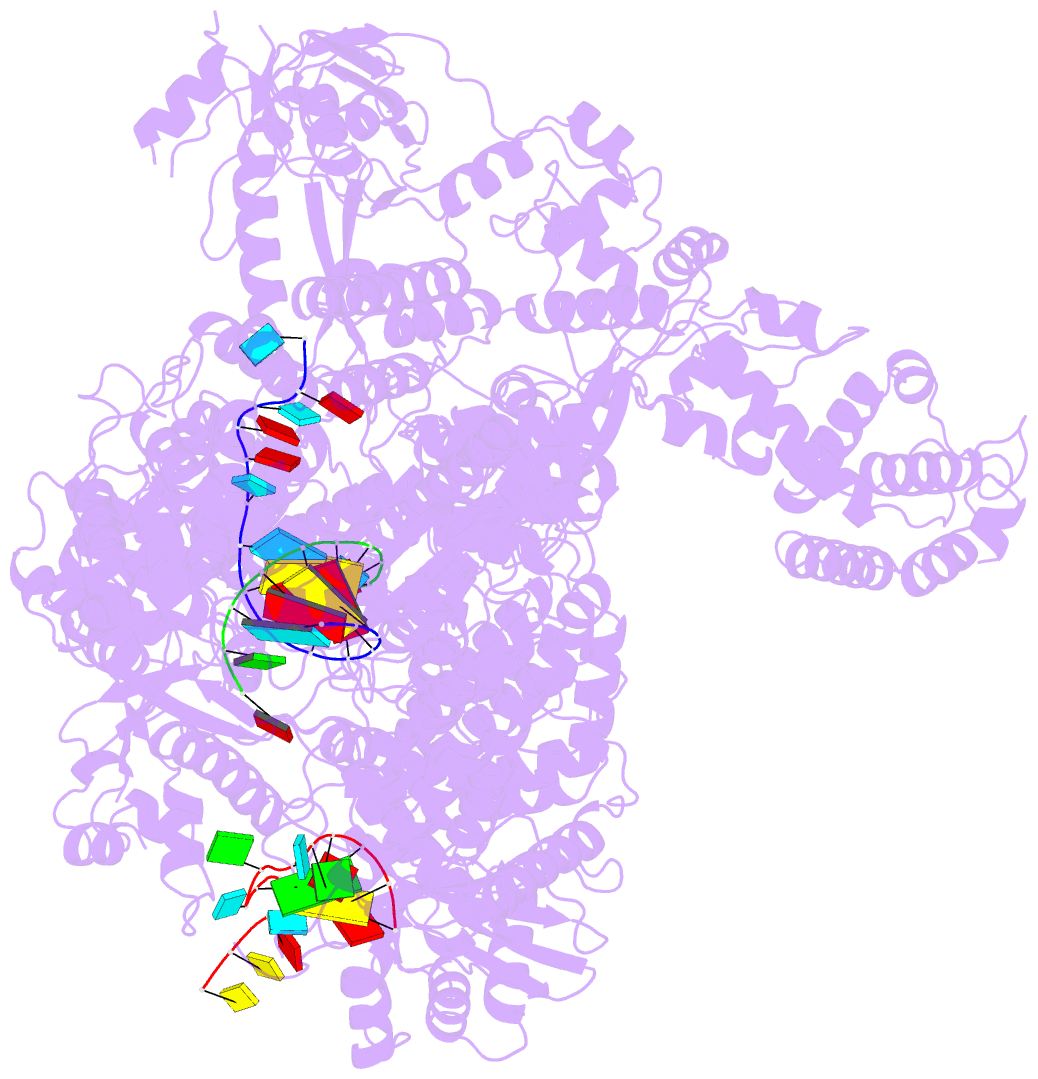
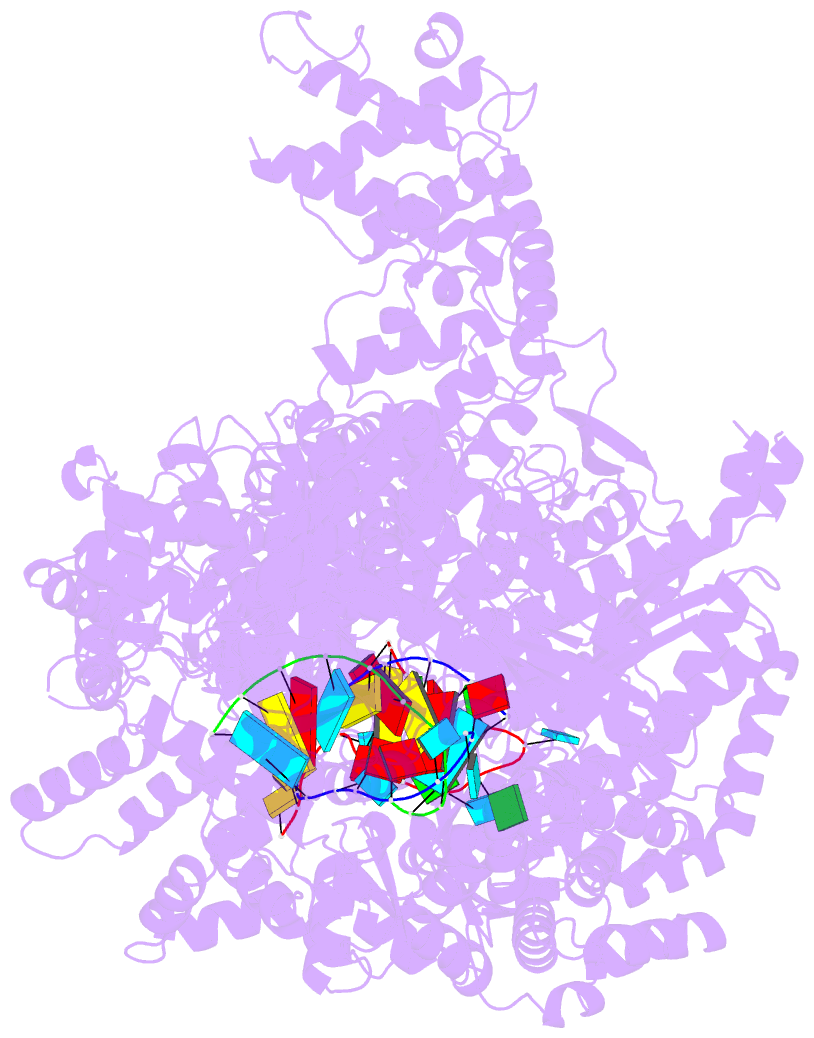
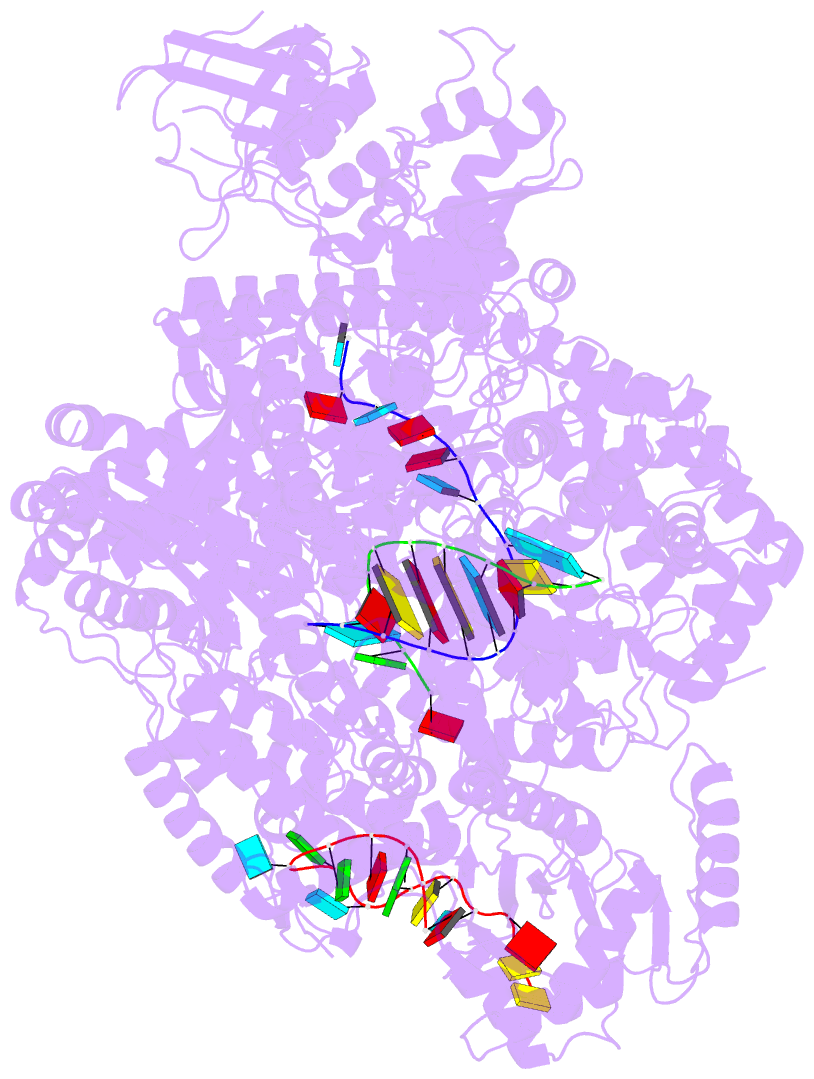
- La crosse virus polymerase at transcription early-elongation stage
- Reference
- Arragain B, Durieux Trouilleton Q, Baudin F, Provaznik J, Azevedo N, Cusack S, Schoehn G, Malet H (2022): "Structural snapshots of La Crosse virus polymerase reveal the mechanisms underlying Peribunyaviridae replication and transcription." Nat Commun, 13, 902. doi: 10.1038/s41467-022-28428-z.
- Abstract
- Segmented negative-strand RNA bunyaviruses encode a multi-functional polymerase that performs genome replication and transcription. Here, we establish conditions for in vitro activity of La Crosse virus polymerase and visualize its conformational dynamics by cryo-electron microscopy, unveiling the precise molecular mechanics underlying its essential activities. We find that replication initiation is coupled to distal duplex promoter formation, endonuclease movement, prime-and-realign loop extension and closure of the polymerase core that direct the template towards the active site. Transcription initiation depends on C-terminal region closure and endonuclease movements that prompt primer cleavage prior to primer entry in the active site. Product realignment after priming, observed in replication and transcription, is triggered by the prime-and-realign loop. Switch to elongation results in polymerase reorganization and core region opening to facilitate template-product duplex formation in the active site cavity. The uncovered detailed mechanics should be helpful for the future design of antivirals counteracting bunyaviral life threatening pathogens.