Summary information and primary citation
- PDB-id
- 7otq; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- cryo-EM (4.8 Å)
- Summary
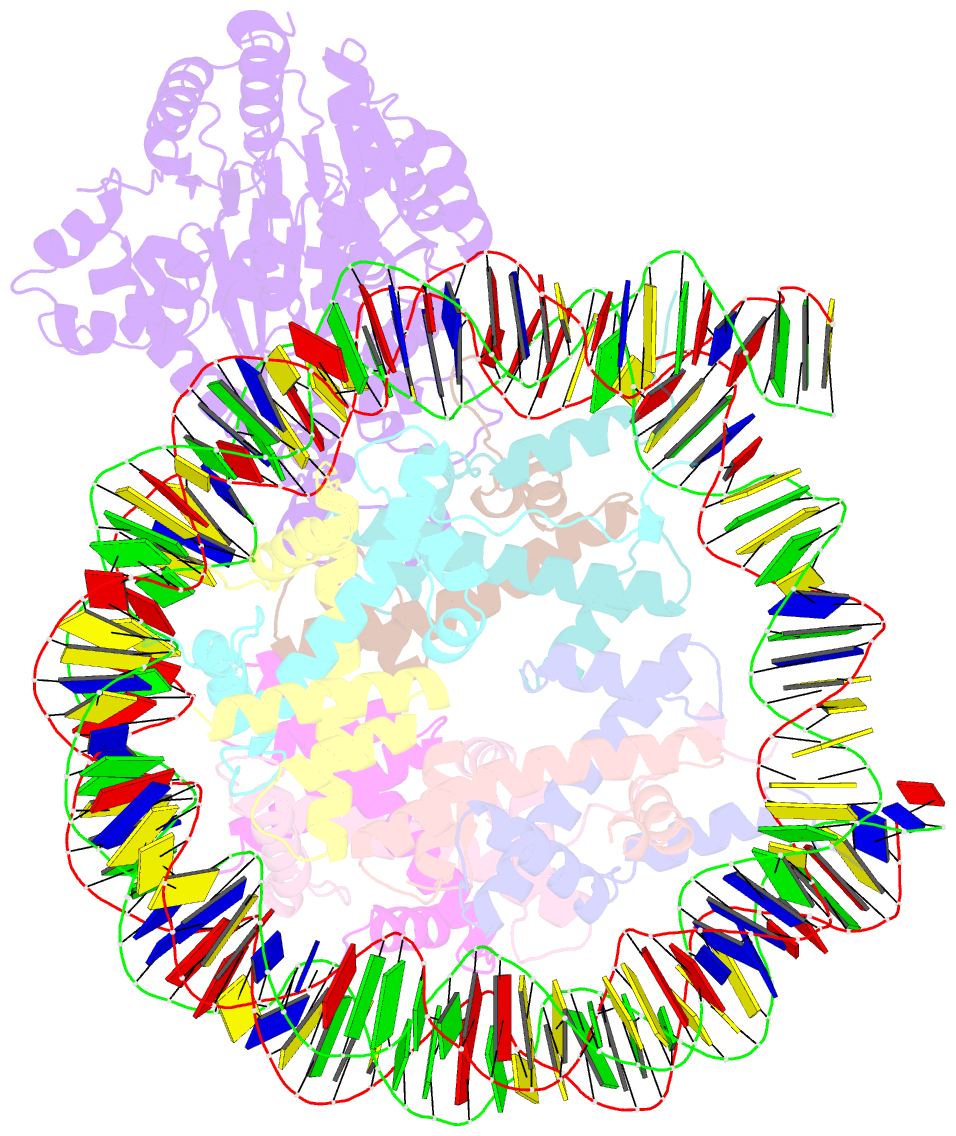
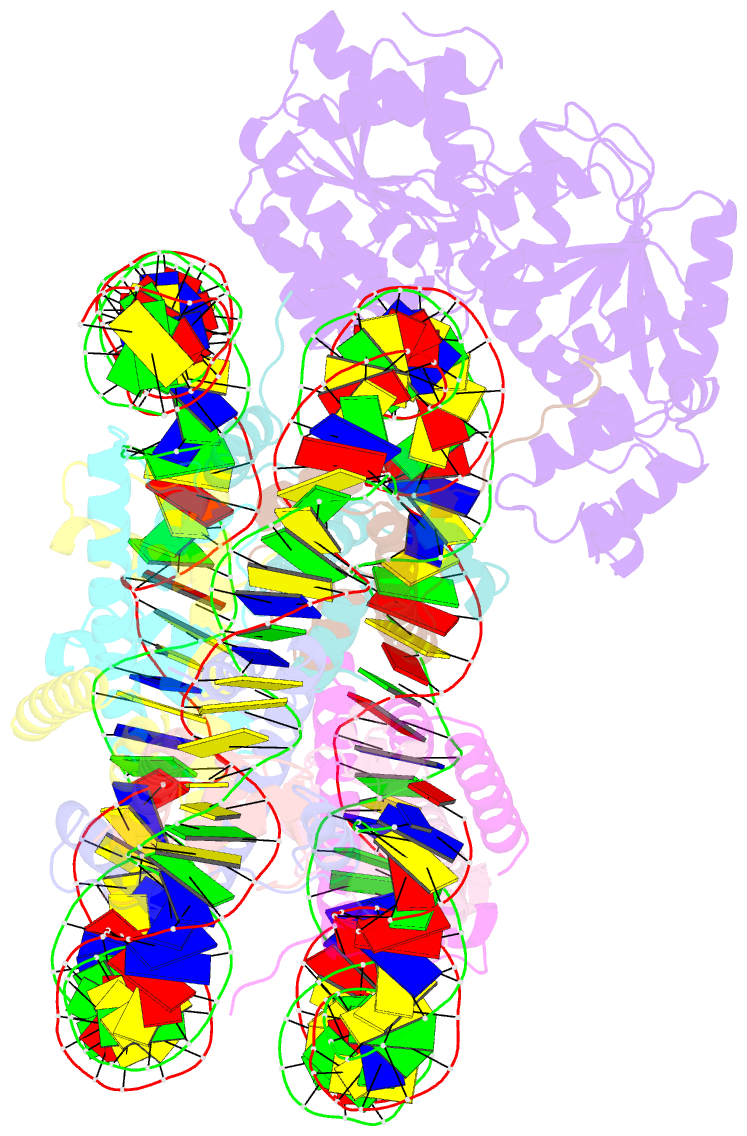
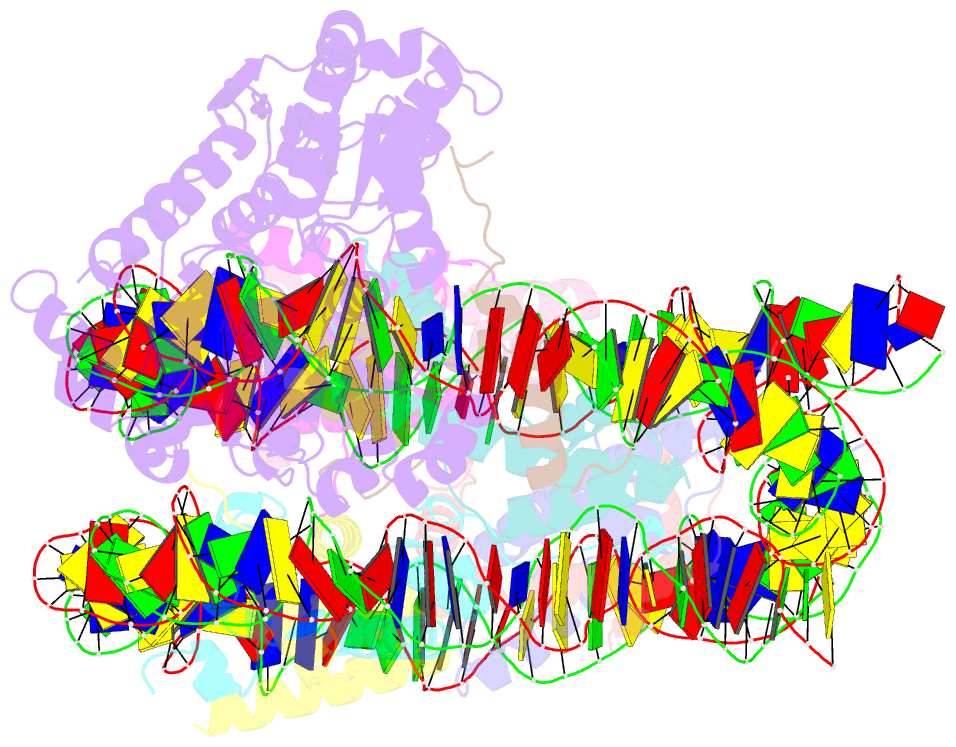
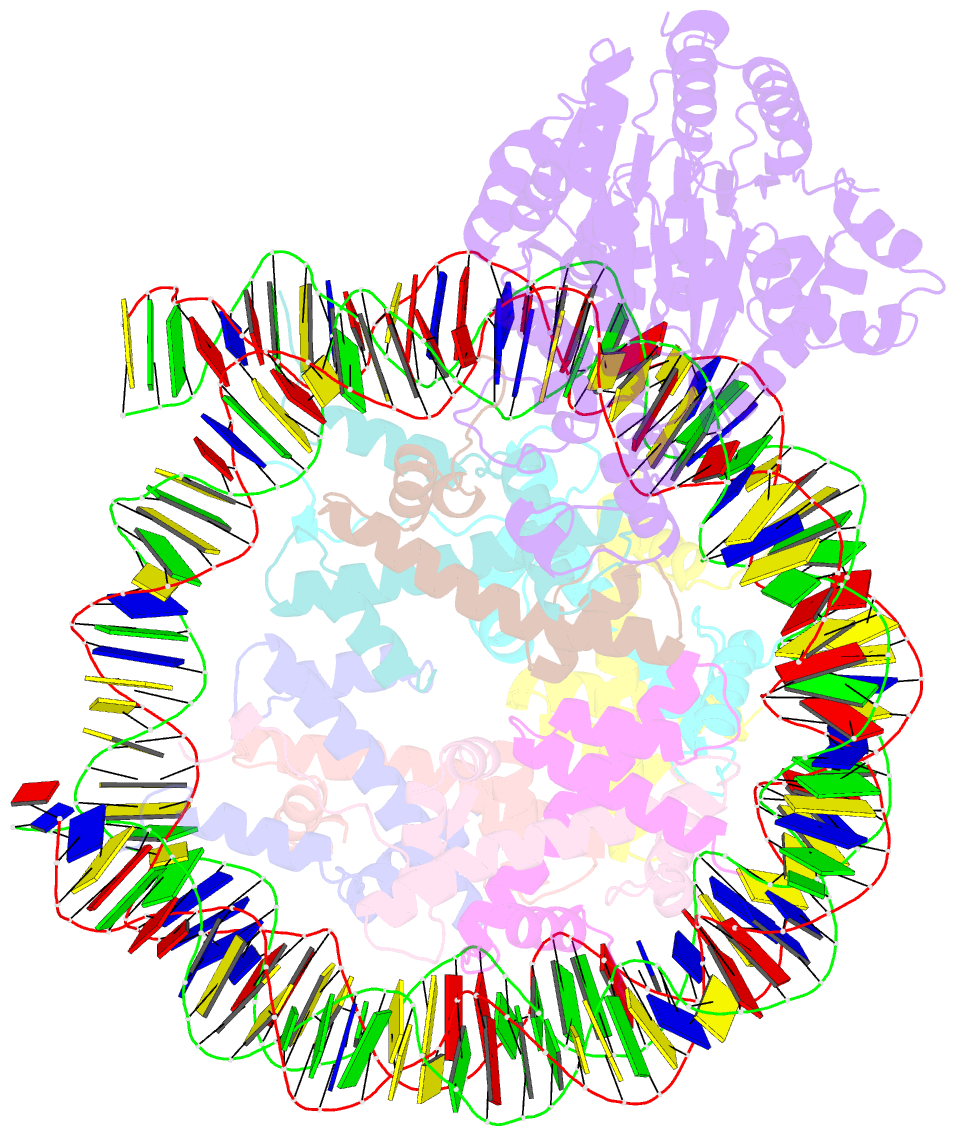
- cryo-EM structure of alc1-chd1l bound to a parylated nucleosome
- Reference
- Bacic L, Gaullier G, Sabantsev A, Lehmann LC, Brackmann K, Dimakou D, Halic M, Hewitt G, Boulton S, Deindl S, Workman JL (2021): "Structure and dynamics of the chromatin remodeler ALC1 bound to a PARylated nucleosome." Elife, 10. doi: 10.7554/eLife.71420.
- Abstract
- The chromatin remodeler ALC1 is recruited to and activated by DNA damage-induced poly(ADP-ribose) (PAR) chains deposited by PARP1/PARP2/HPF1 upon detection of DNA lesions. ALC1 has emerged as a candidate drug target for cancer therapy as its loss confers synthetic lethality in homologous recombination-deficient cells. However, structure-based drug design and molecular analysis of ALC1 have been hindered by the requirement for PARylation and the highly heterogeneous nature of this post-translational modification. Here, we reconstituted an ALC1 and PARylated nucleosome complex modified in vitro using PARP2 and HPF1. This complex was amenable to cryo-EM structure determination without cross-linking, which enabled visualization of several intermediate states of ALC1 from the recognition of the PARylated nucleosome to the tight binding and activation of the remodeler. Functional biochemical assays with PARylated nucleosomes highlight the importance of nucleosomal epitopes for productive remodeling and suggest that ALC1 preferentially slides nucleosomes away from DNA breaks.