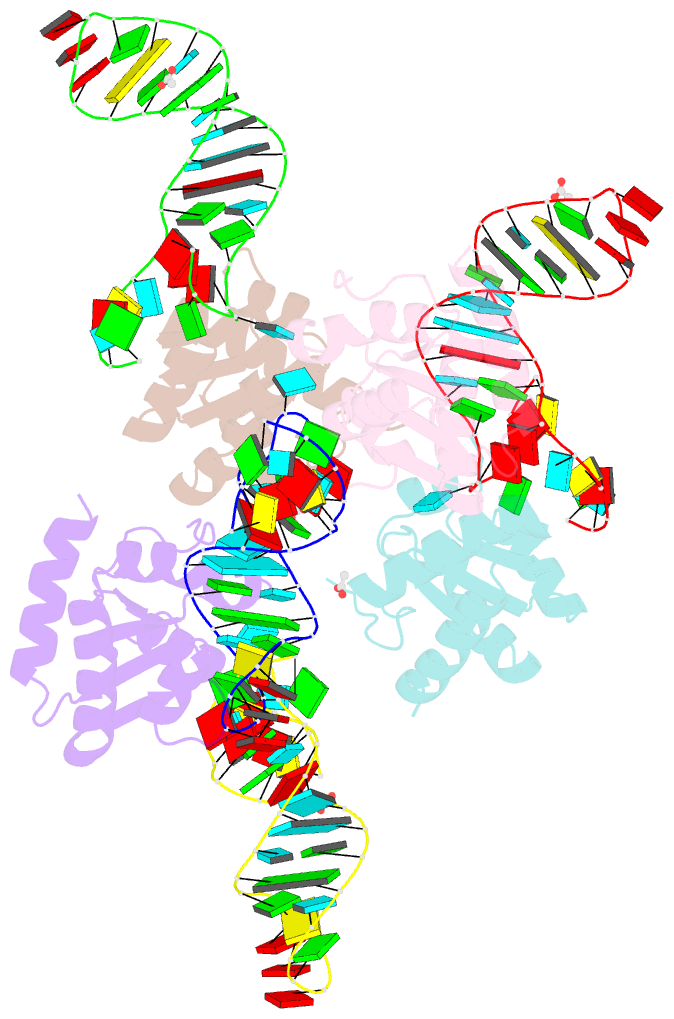
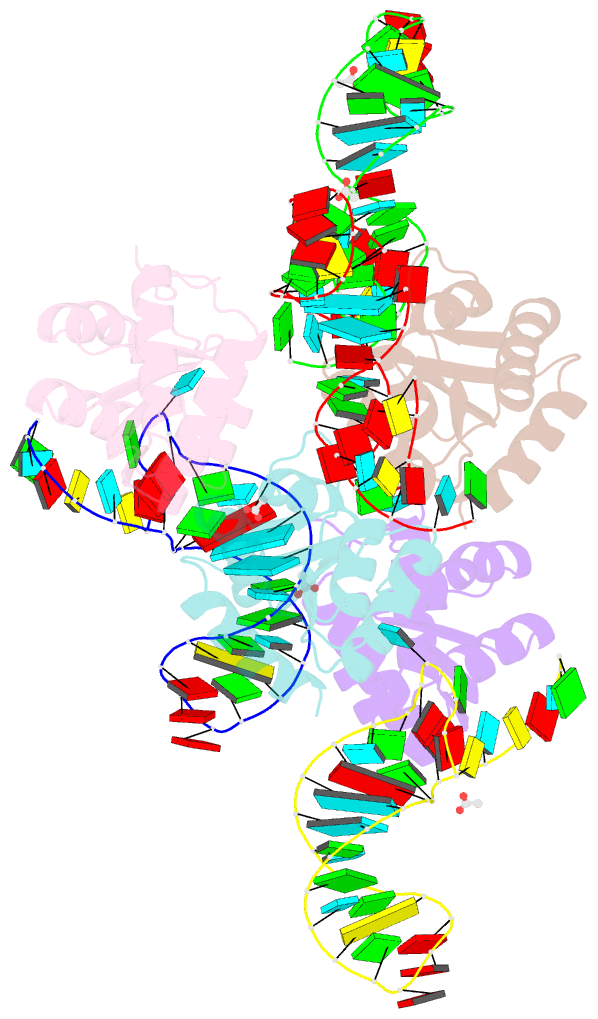
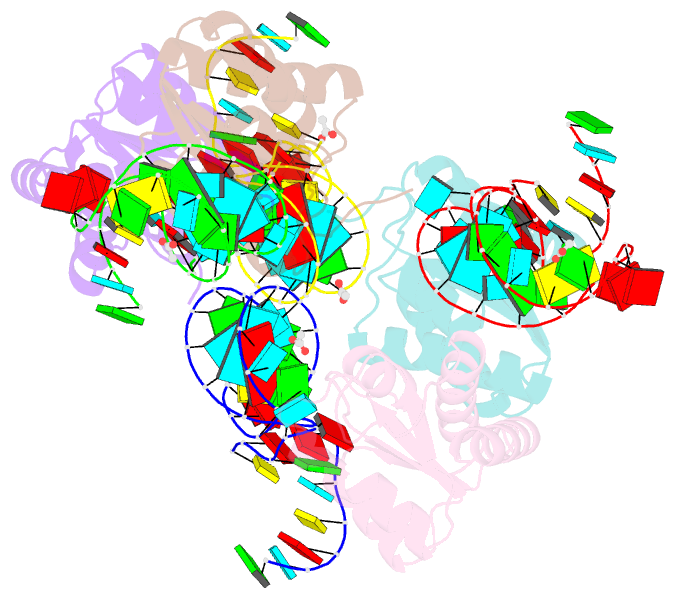
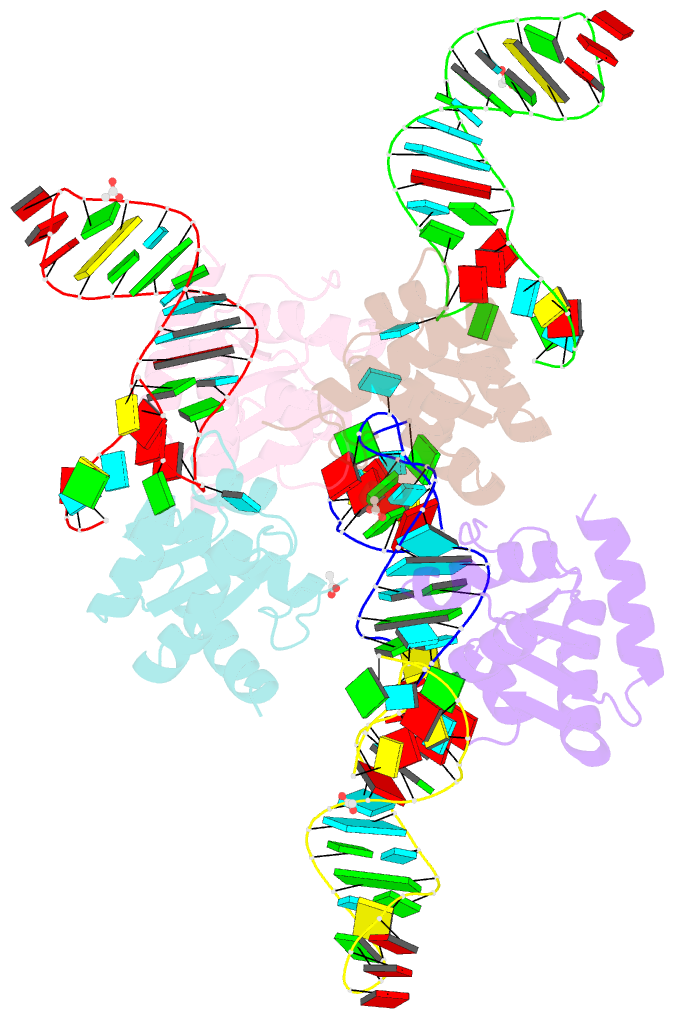
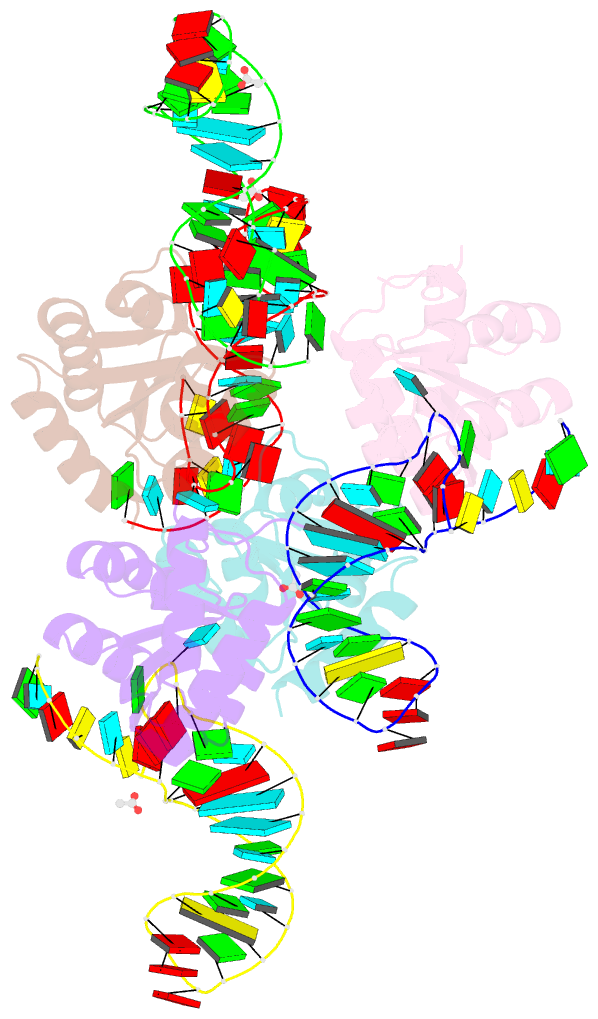
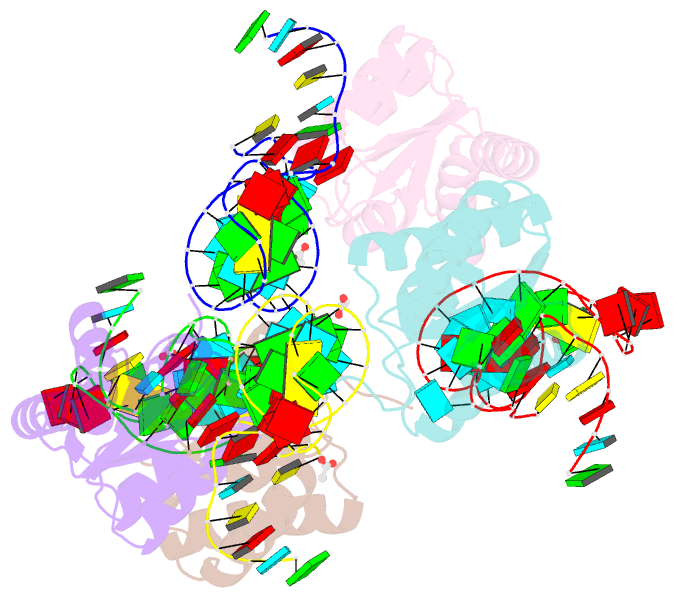
Summary information and primary citation
- PDB-id
- 7ozq; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ribosomal protein
- Method
- X-ray (1.91 Å)
- Summary
- Crystal structure of archaeal l7ae bound to eukaryotic kink-loop
- Reference
- Hofler S, Lukat P, Blankenfeldt W, Carlomagno T (2021): "Eukaryotic Box C/D methylation machinery has two non-symmetric protein assembly sites." Sci Rep, 11, 17561. doi: 10.1038/s41598-021-97030-y.
- Abstract
- Box C/D ribonucleoprotein complexes are RNA-guided methyltransferases that methylate the ribose 2'-OH of RNA. The central 'guide RNA' has box C and D motifs at its ends, which are crucial for activity. Archaeal guide RNAs have a second box C'/D' motif pair that is also essential for function. This second motif is poorly conserved in eukaryotes and its function is uncertain. Conflicting literature data report that eukaryotic box C'/D' motifs do or do not bind proteins specialized to recognize box C/D-motifs and are or are not important for function. Despite this uncertainty, the architecture of eukaryotic 2'-O-methylation enzymes is thought to be similar to that of their archaeal counterpart. Here, we use biochemistry, X-ray crystallography and mutant analysis to demonstrate the absence of functional box C'/D' motifs in more than 80% of yeast guide RNAs. We conclude that eukaryotic Box C/D RNPs have two non-symmetric protein assembly sites and that their three-dimensional architecture differs from that of archaeal 2'-O-methylation enzymes.