Summary information and primary citation
- PDB-id
- 7q2z; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- cell cycle
- Method
- cryo-EM (3.2 Å)
- Summary
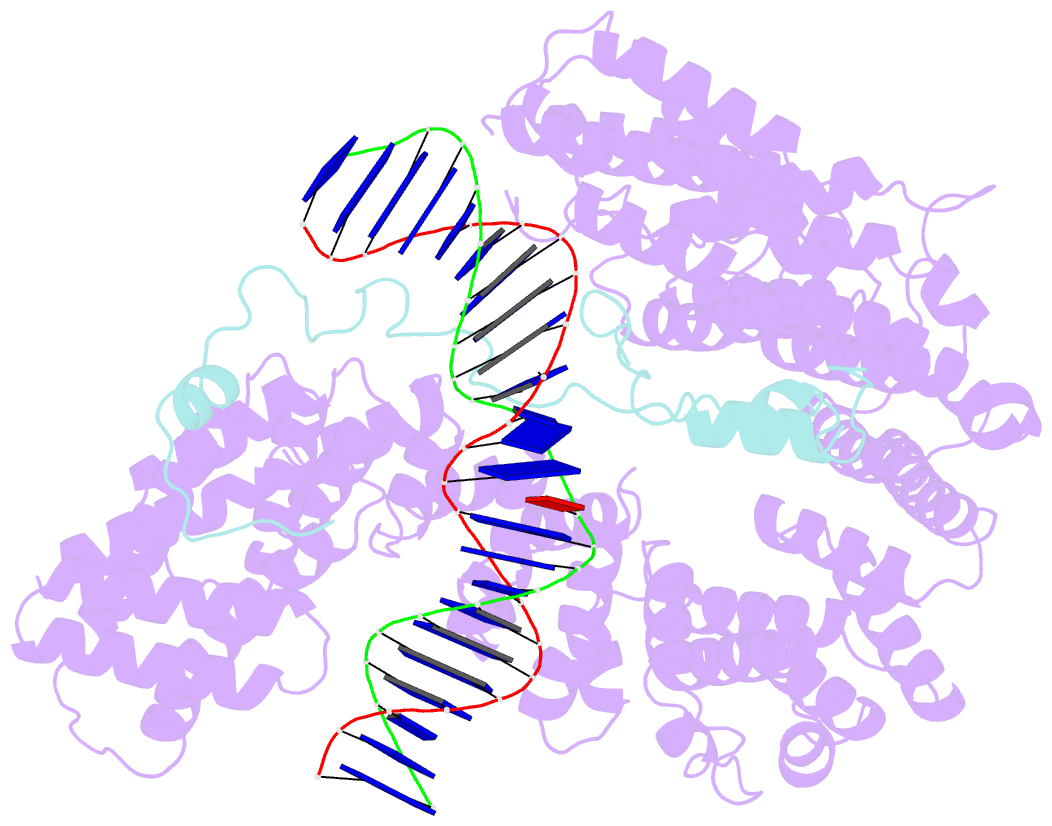
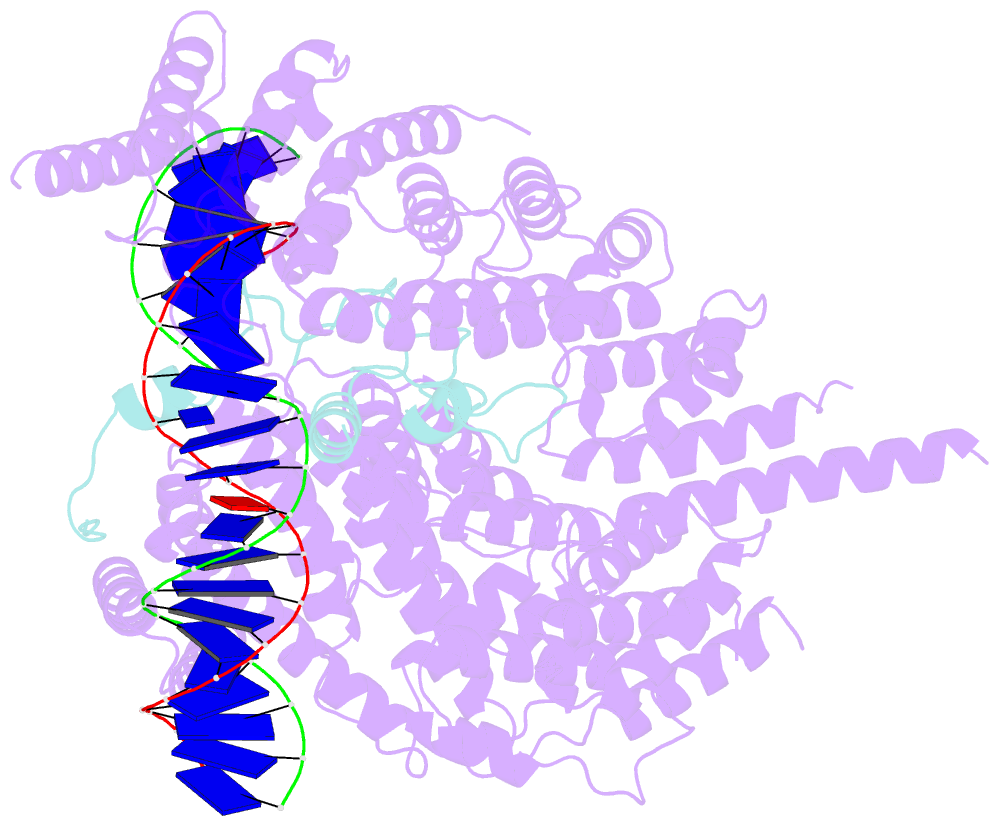
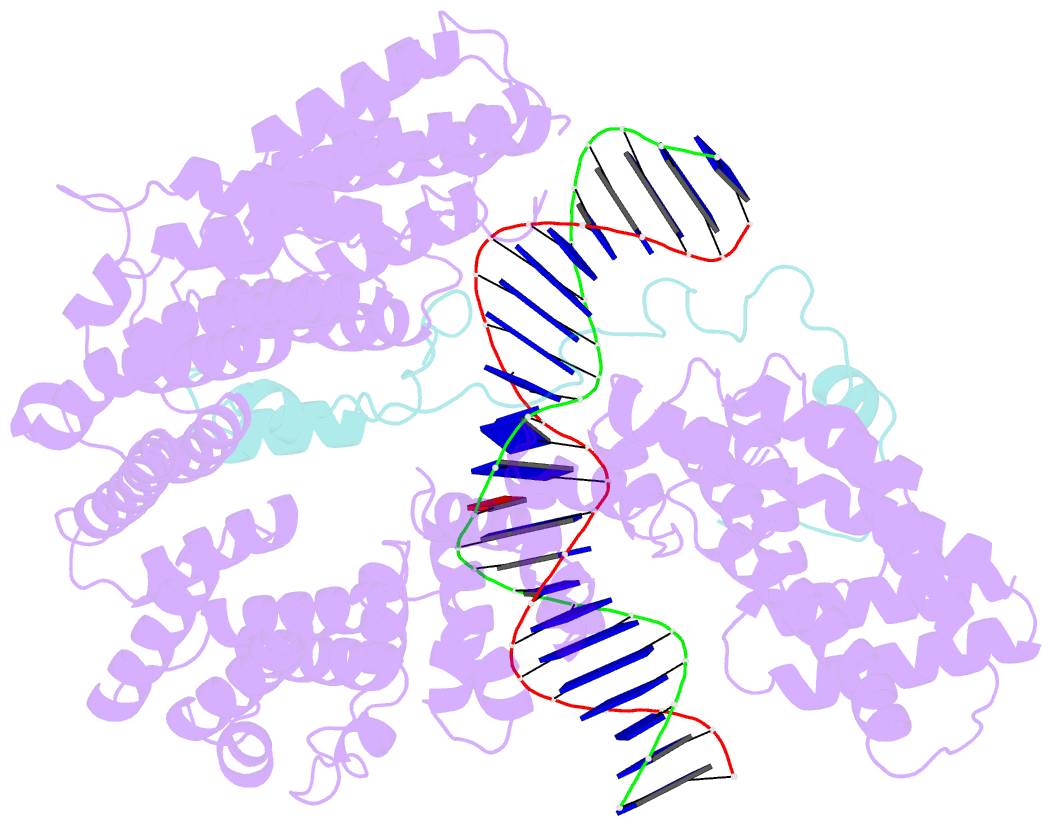
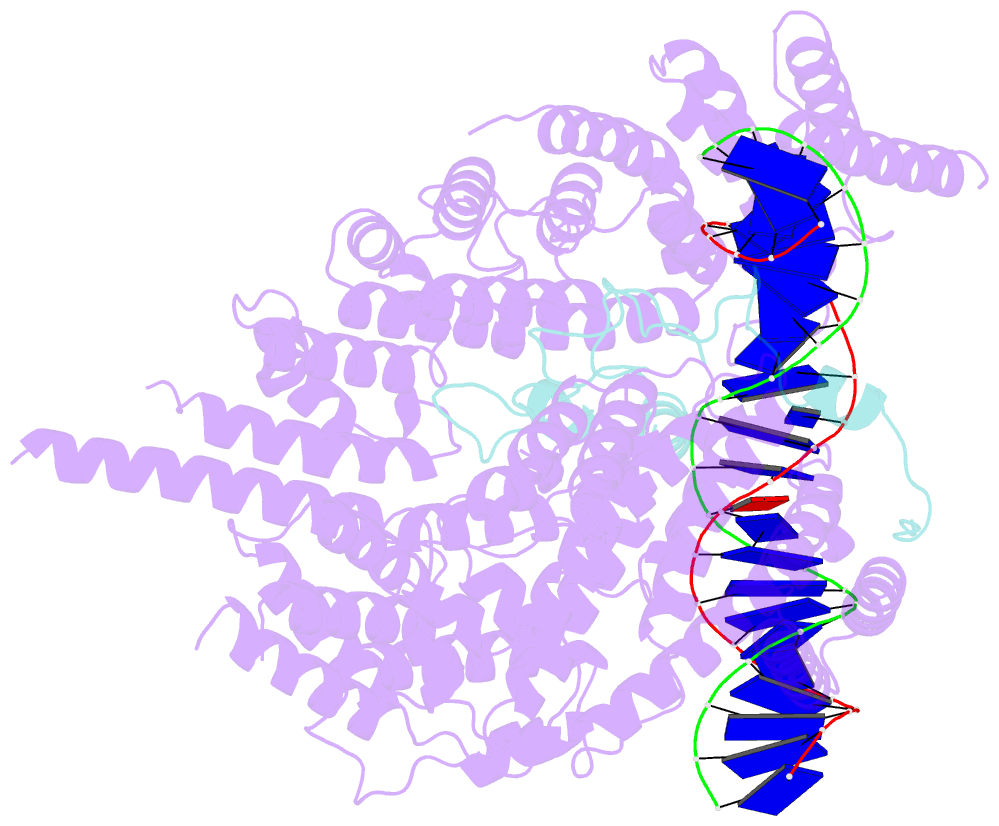
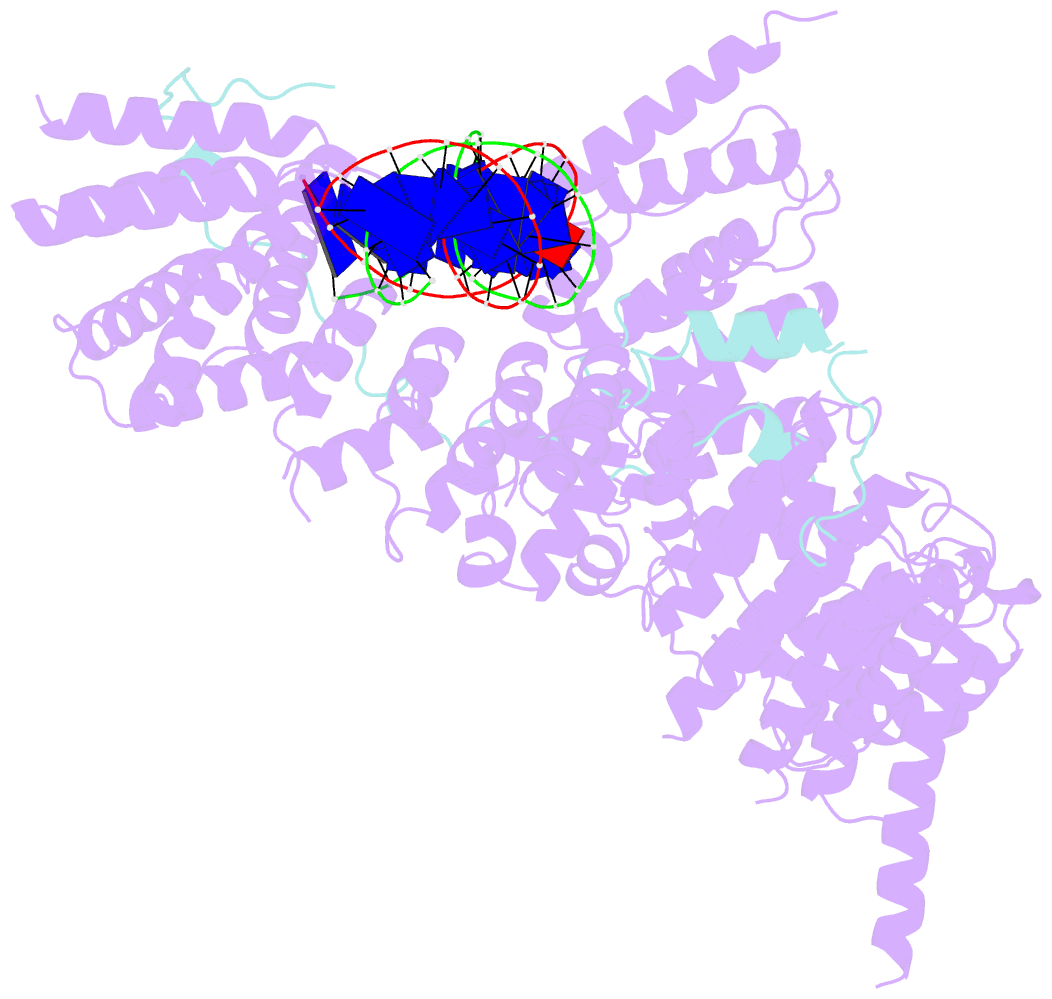
- cryo-EM structure of s.cerevisiae condensin ycg1-brn1-DNA complex
- Reference
- Lee BG, Rhodes J, Lowe J (2022): "Clamping of DNA shuts the condensin neck gate." Proc.Natl.Acad.Sci.USA, 119, e2120006119. doi: 10.1073/pnas.2120006119.
- Abstract
- SignificanceDNA needs to be compacted to fit into nuclei and during cell division, when dense chromatids are formed for their mechanical segregation, a process that depends on the protein complex condensin. It forms and enlarges loops in DNA through loop extrusion. Our work resolves the atomic structure of a DNA-bound state of condensin in which ATP has not been hydrolyzed. The DNA is clamped within a compartment that has been reported previously in other structural maintenance of chromosomes (SMC) complexes, including Rad50, cohesin, and MukBEF. With the caveat of important differences, it means that all SMC complexes cycle through at least some similar states and undergo similar conformational changes in their head modules, while hydrolyzing ATP and translocating DNA.