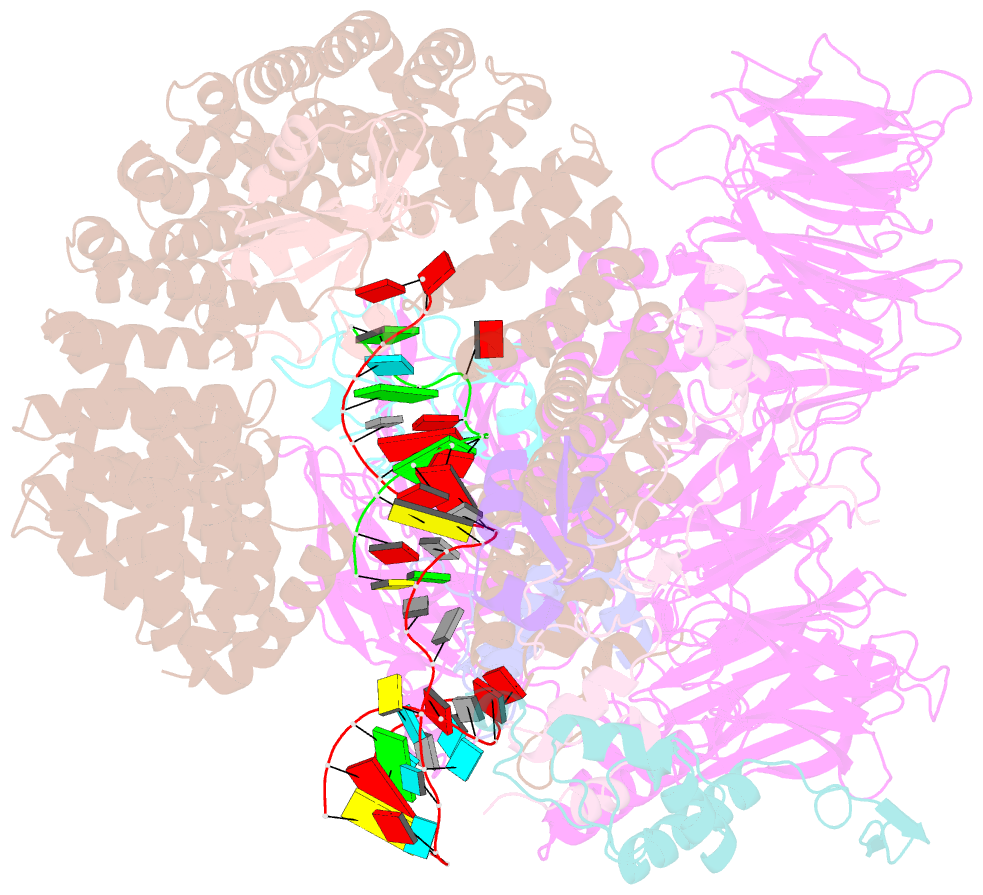
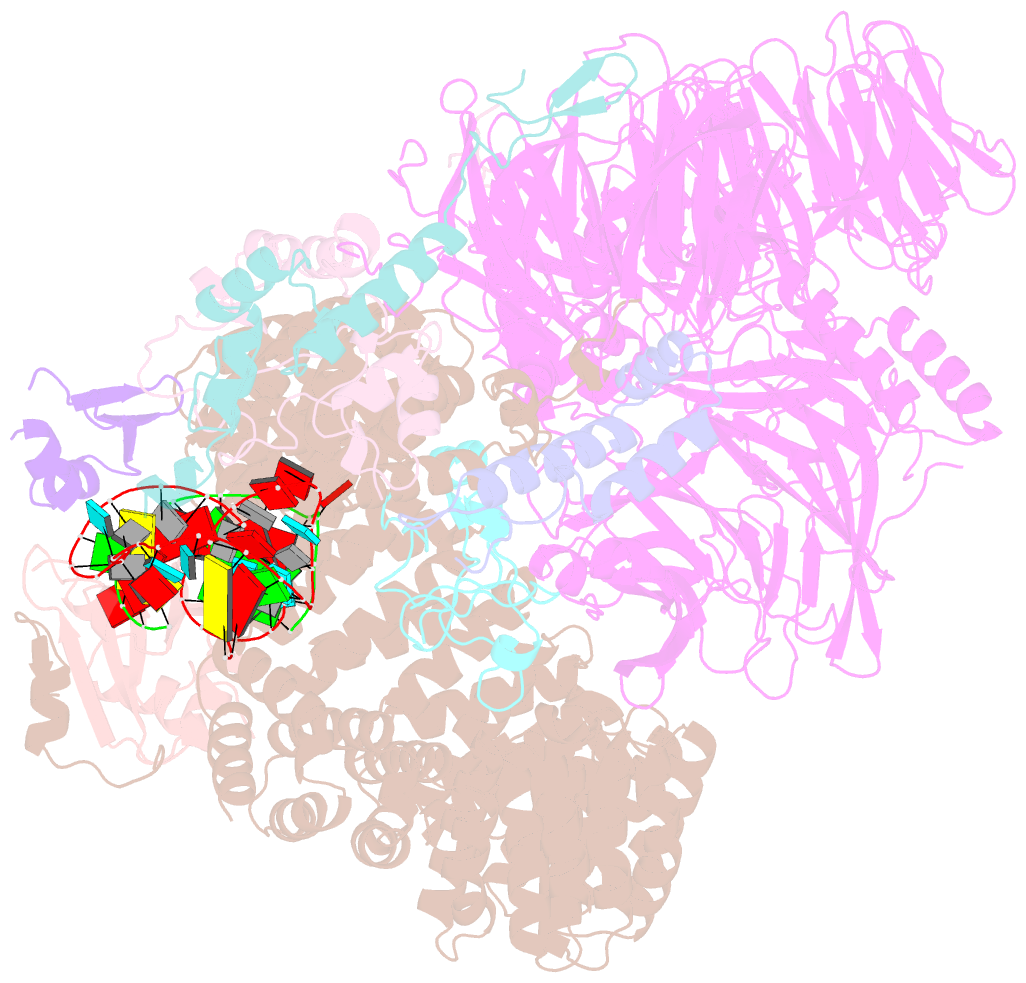
Summary information and primary citation
- PDB-id
- 7q4o; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- nuclear protein
- Method
- cryo-EM (2.1 Å)
- Summary
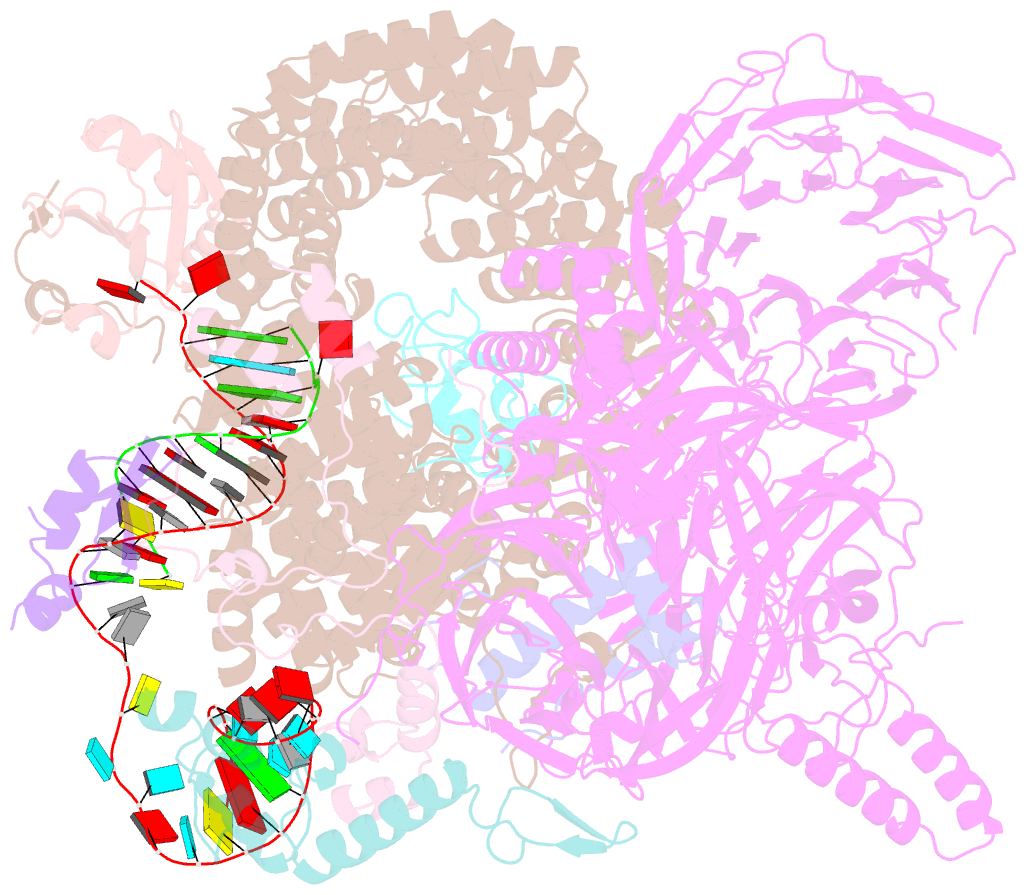
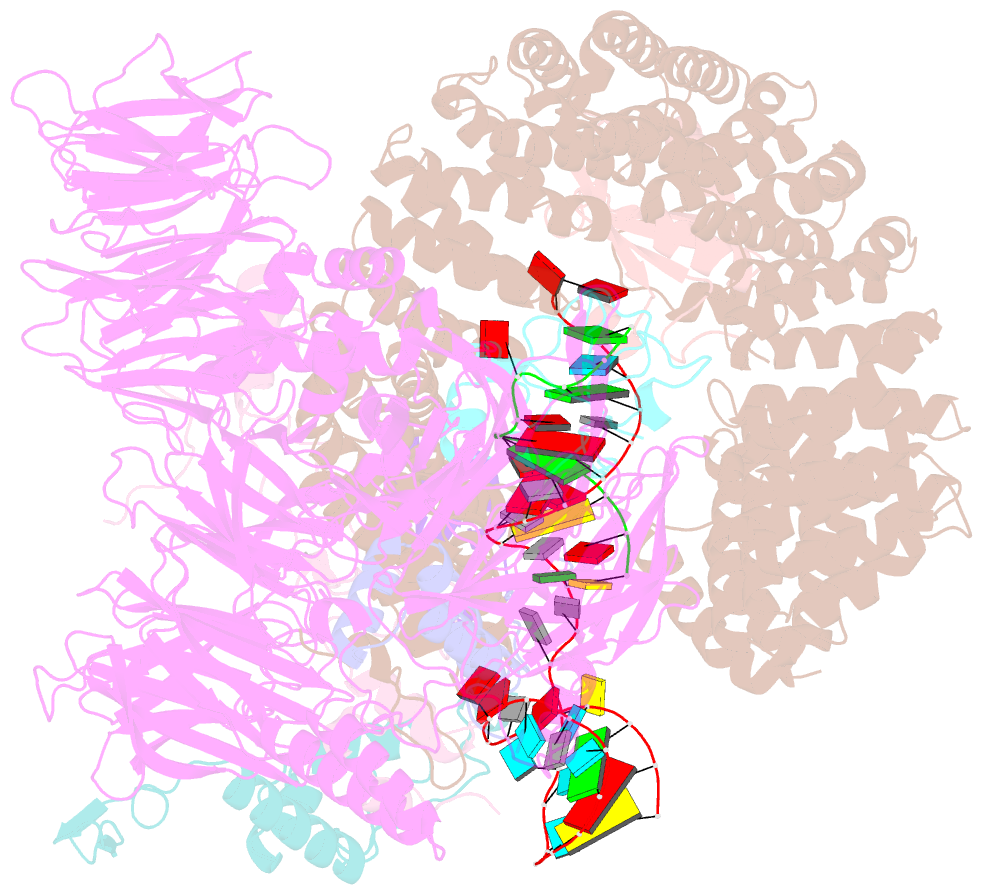
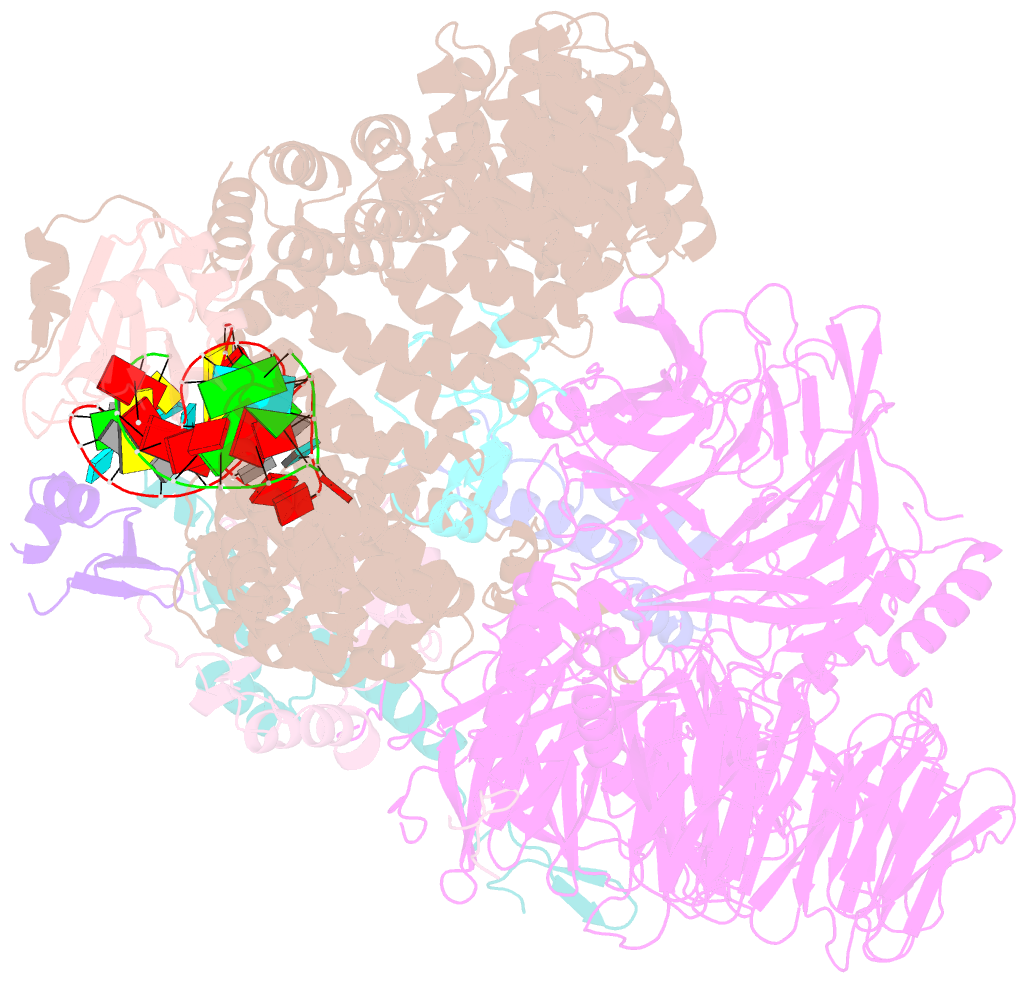
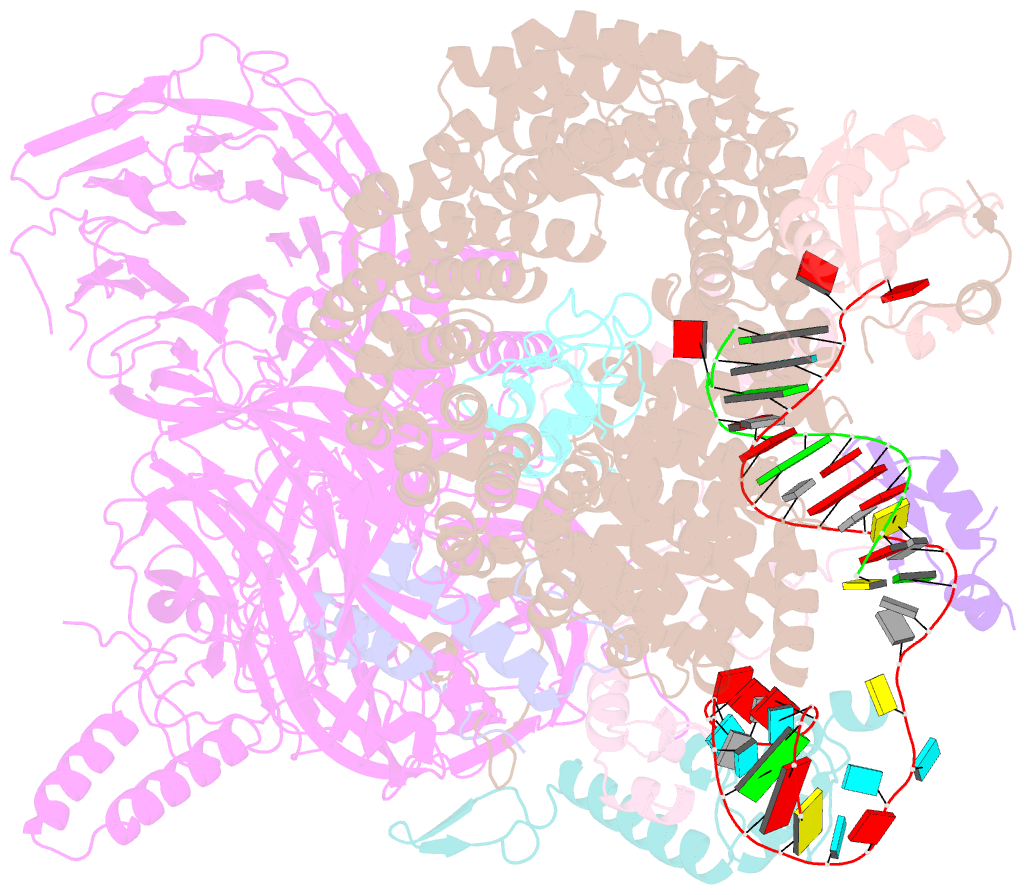
- Substrate-bound a-like u2 snrnp
- Reference
- Tholen J, Razew M, Weis F, Galej WP (2022): "Structural basis of branch site recognition by the human spliceosome." Science, 375, 50-57. doi: 10.1126/science.abm4245.
- Abstract
- Recognition of the intron branch site (BS) by the U2 small nuclear ribonucleoprotein (snRNP) is a critical event during spliceosome assembly. In mammals, BS sequences are poorly conserved, and unambiguous intron recognition cannot be achieved solely through a base-pairing mechanism. We isolated human 17S U2 snRNP and reconstituted in vitro its adenosine 5´-triphosphate (ATP)–dependent remodeling and binding to the pre–messenger RNA substrate. We determined a series of high-resolution (2.0 to 2.2 angstrom) structures providing snapshots of the BS selection process. The substrate-bound U2 snRNP shows that SF3B6 stabilizes the BS:U2 snRNA duplex, which could aid binding of introns with poor sequence complementarity. ATP-dependent remodeling uncoupled from substrate binding captures U2 snRNA in a conformation that competes with BS recognition, providing a selection mechanism based on branch helix stability.