Summary information and primary citation
- PDB-id
- 7q5b; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- cryo-EM (3.98 Å)
- Summary
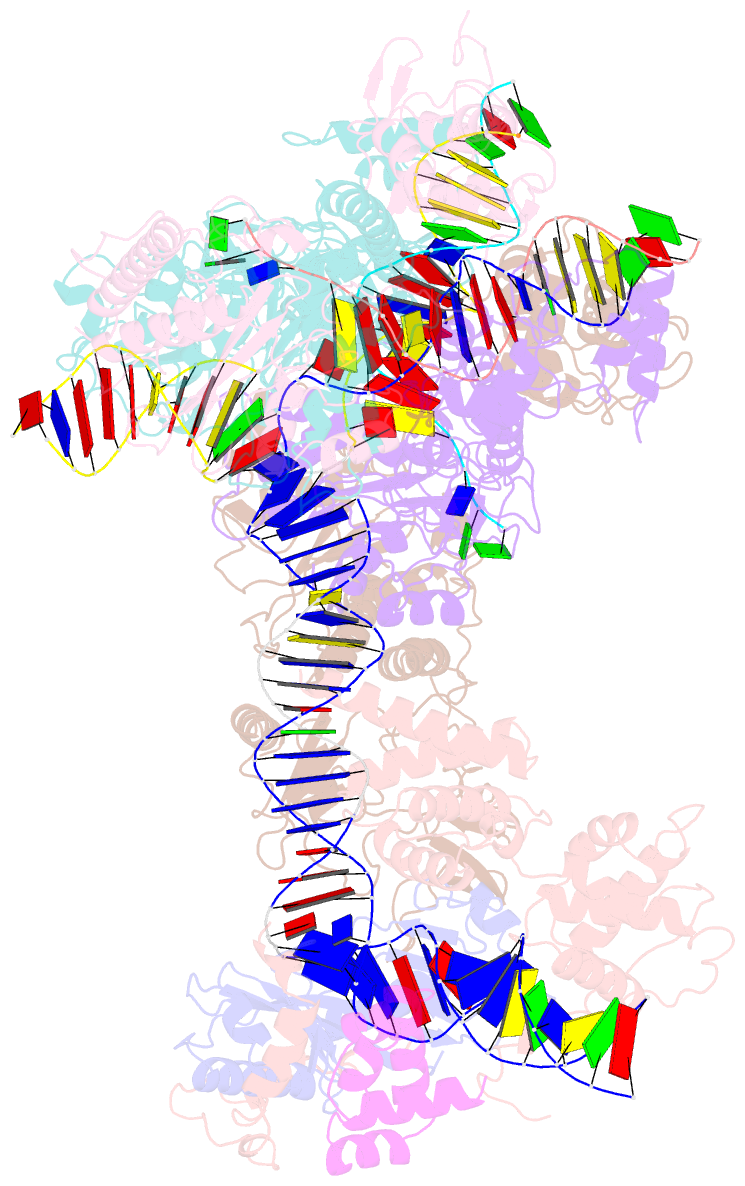
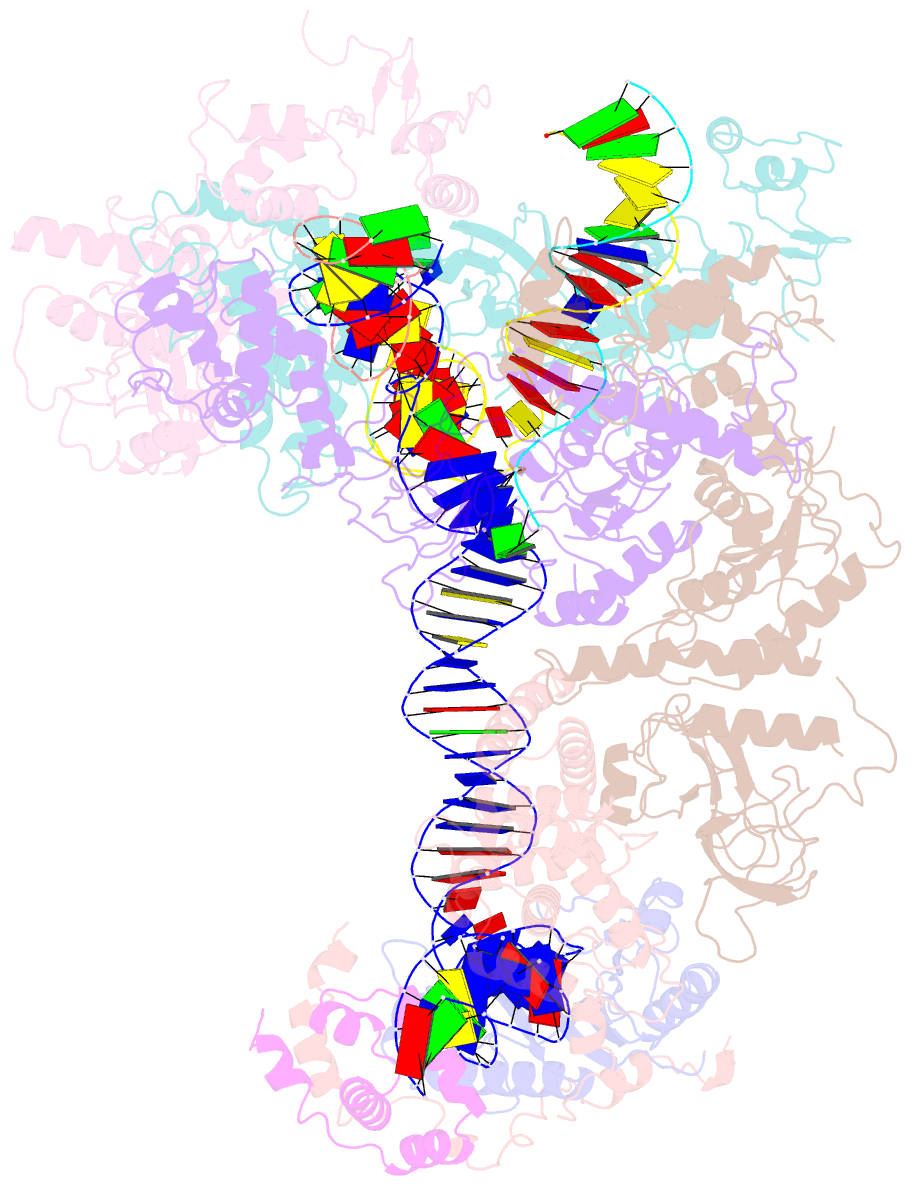
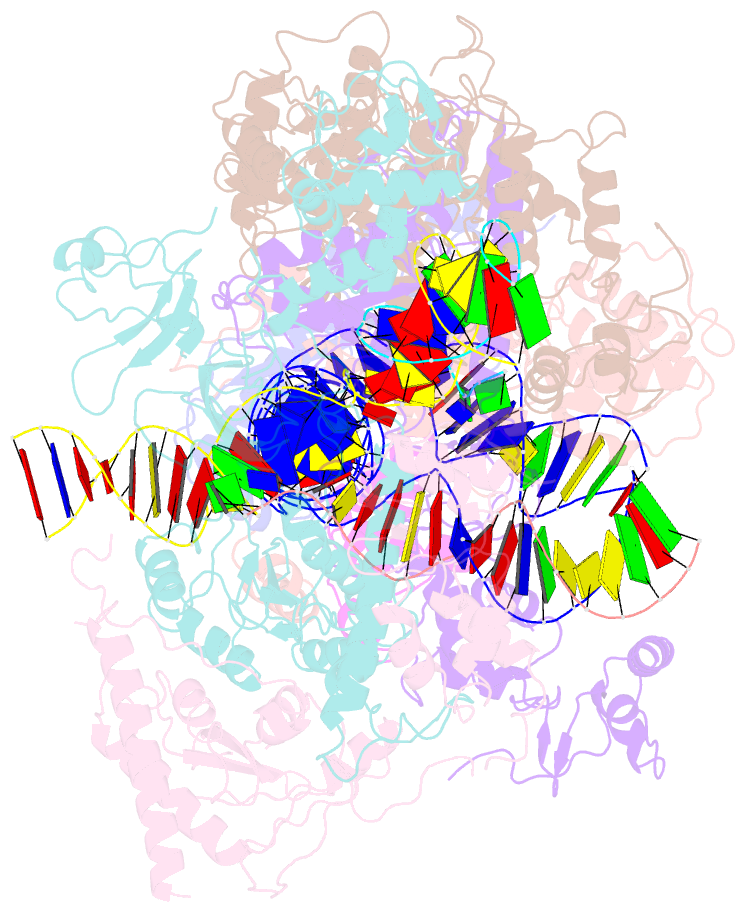
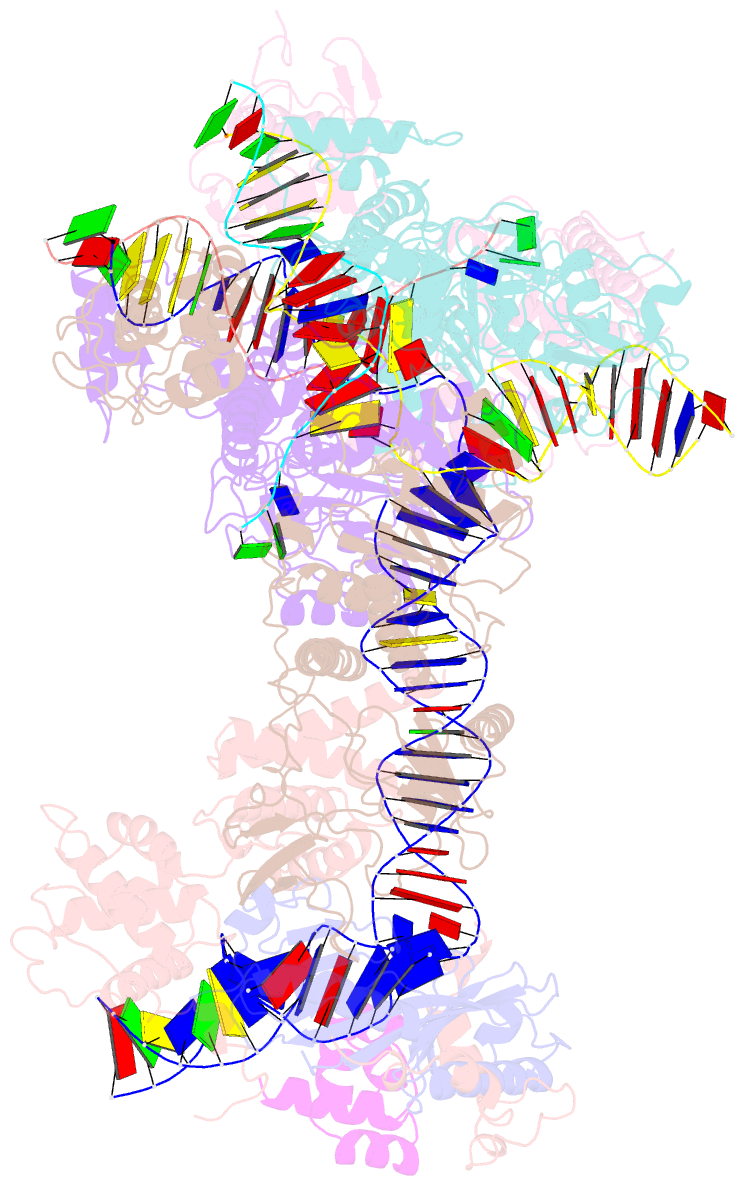
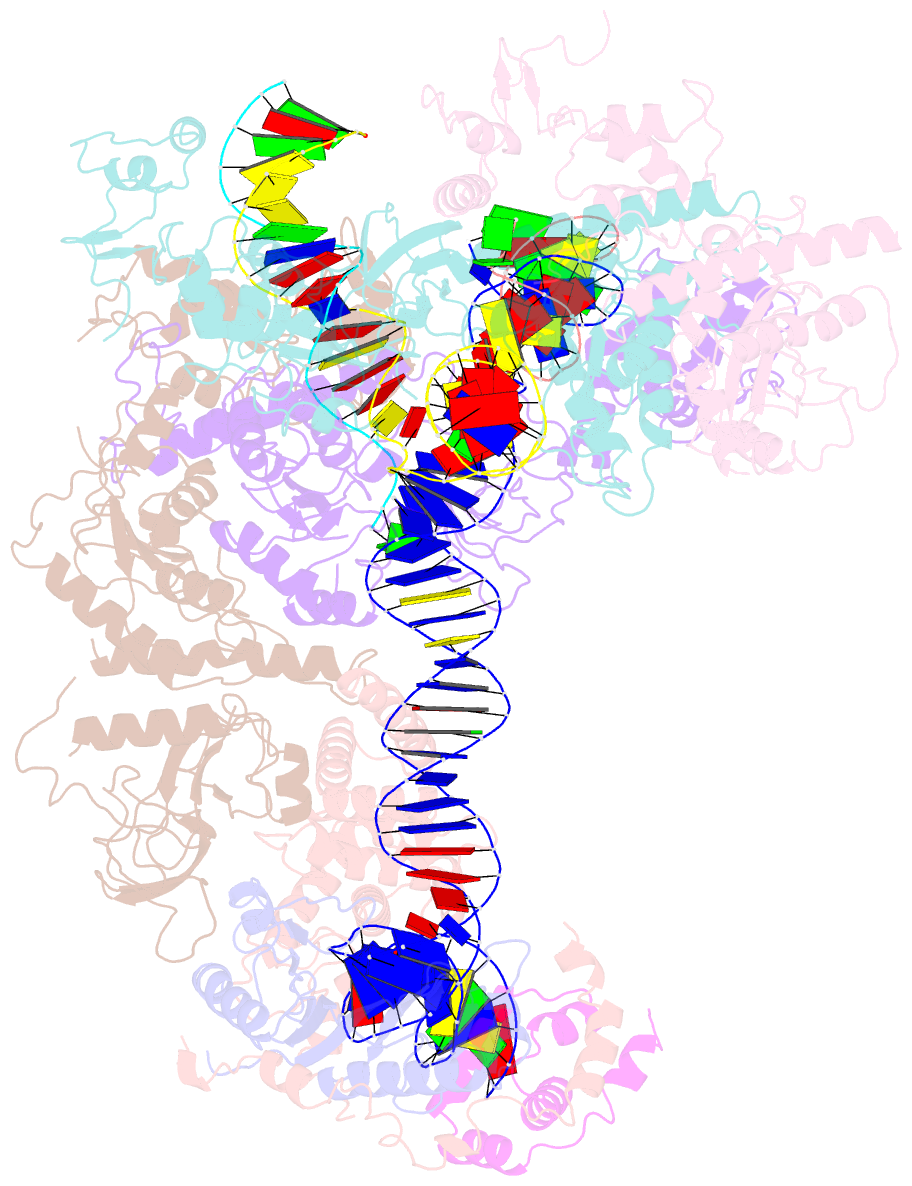
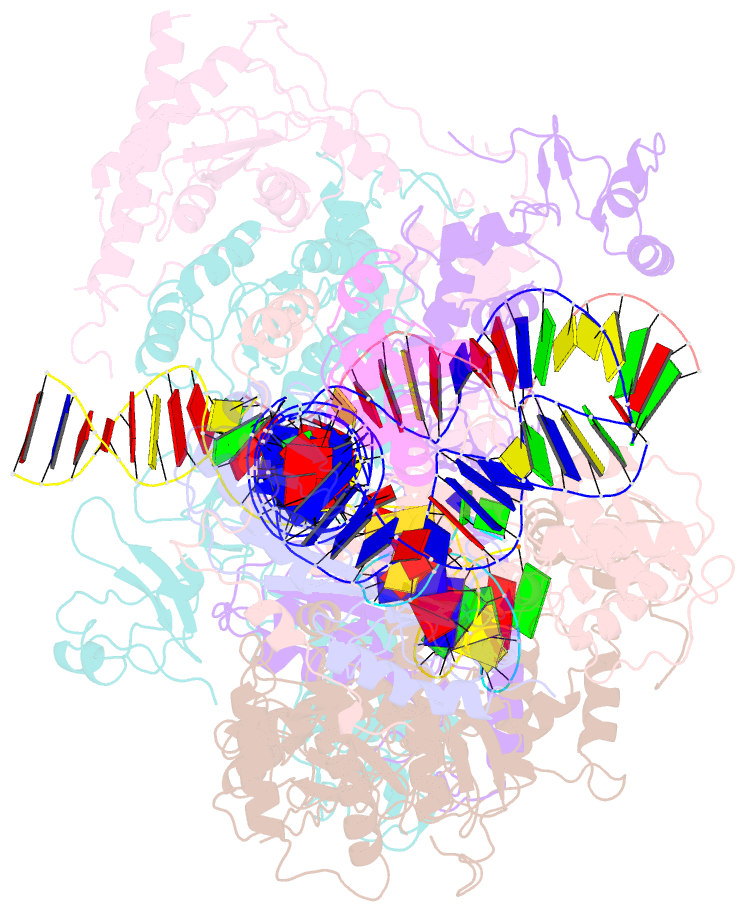
- cryo-EM structure of ty3 retrotransposon targeting a tfiiib-bound trna gene
- Reference
- Abascal-Palacios G, Jochem L, Pla-Prats C, Beuron F, Vannini A (2021): "Structural basis of Ty3 retrotransposon integration at RNA Polymerase III-transcribed genes." Nat Commun, 12, 6992. doi: 10.1038/s41467-021-27338-w.
- Abstract
- Retrotransposons are endogenous elements that have the ability to mobilise their DNA between different locations in the host genome. The Ty3 retrotransposon integrates with an exquisite specificity in a narrow window upstream of RNA Polymerase (Pol) III-transcribed genes, representing a paradigm for harmless targeted integration. Here we present the cryo-EM reconstruction at 4.0 Å of an active Ty3 strand transfer complex bound to TFIIIB transcription factor and a tRNA gene. The structure unravels the molecular mechanisms underlying Ty3 targeting specificity at Pol III-transcribed genes and sheds light into the architecture of retrotransposon machinery during integration. Ty3 intasome contacts a region of TBP, a subunit of TFIIIB, which is blocked by NC2 transcription regulator in RNA Pol II-transcribed genes. A newly-identified chromodomain on Ty3 integrase interacts with TFIIIB and the tRNA gene, defining with extreme precision the integration site position.