Summary information and primary citation
- PDB-id
- 7qfw; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- cryo-EM (3.86 Å)
- Summary
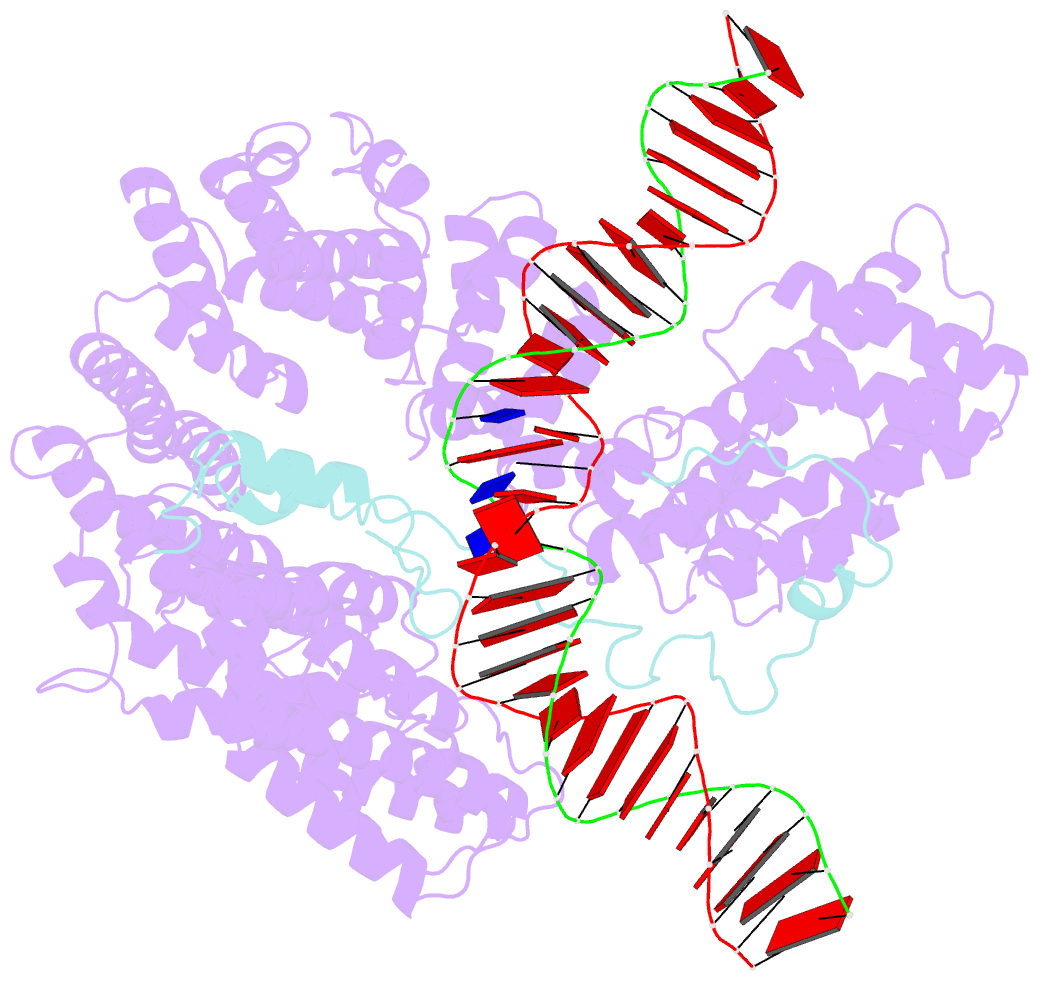
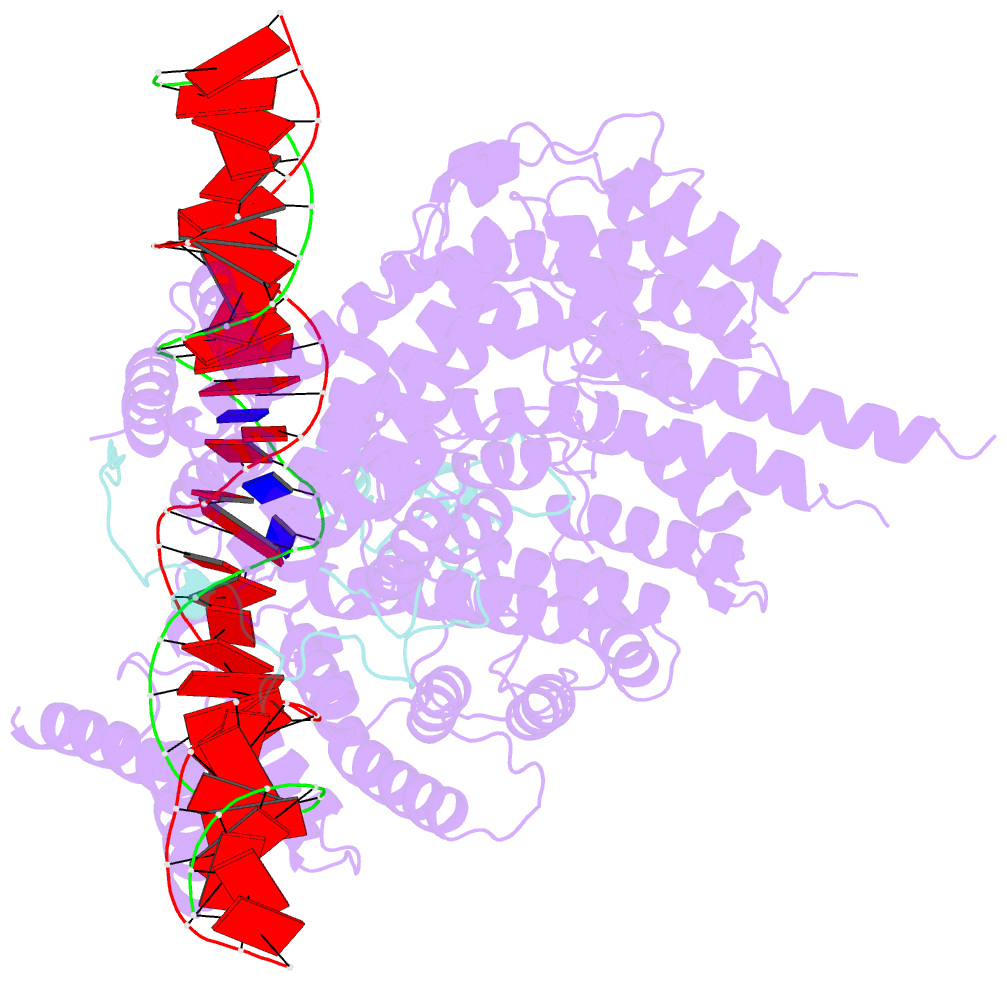
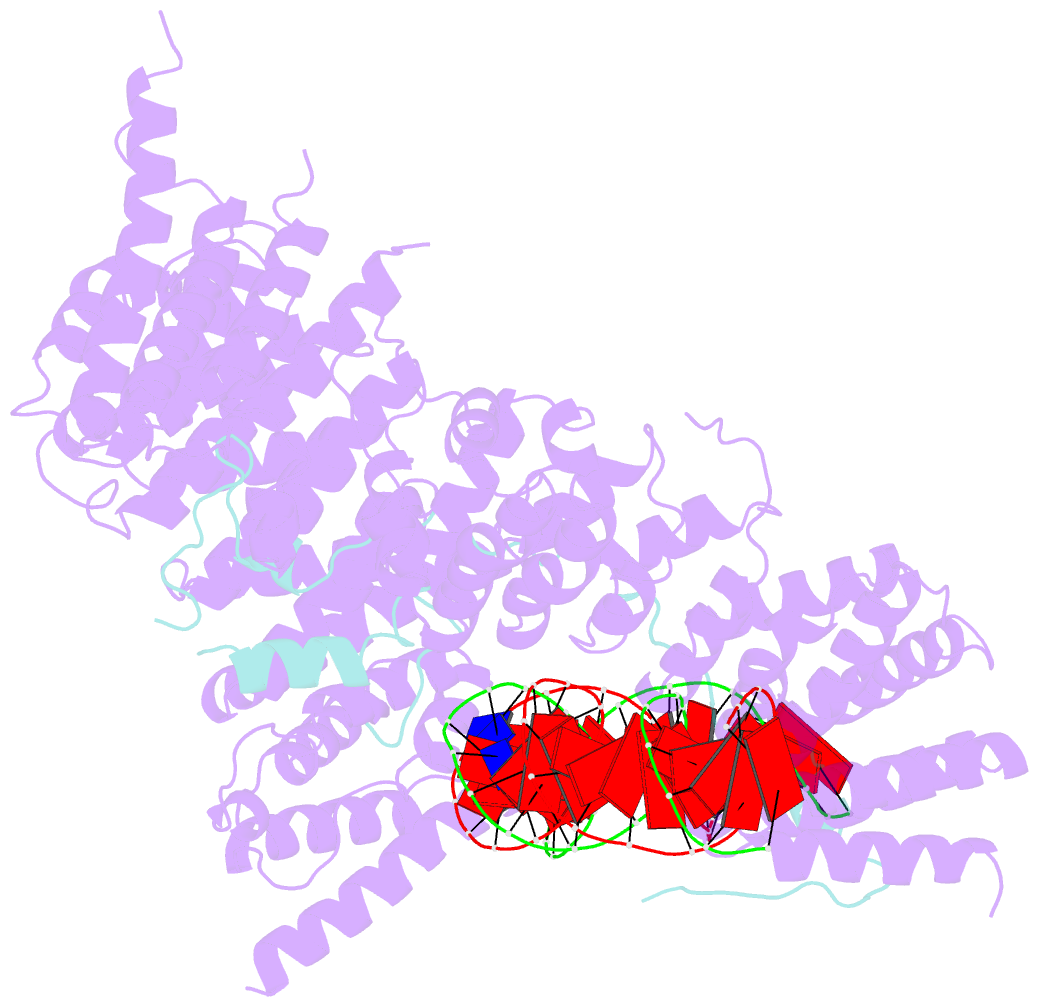
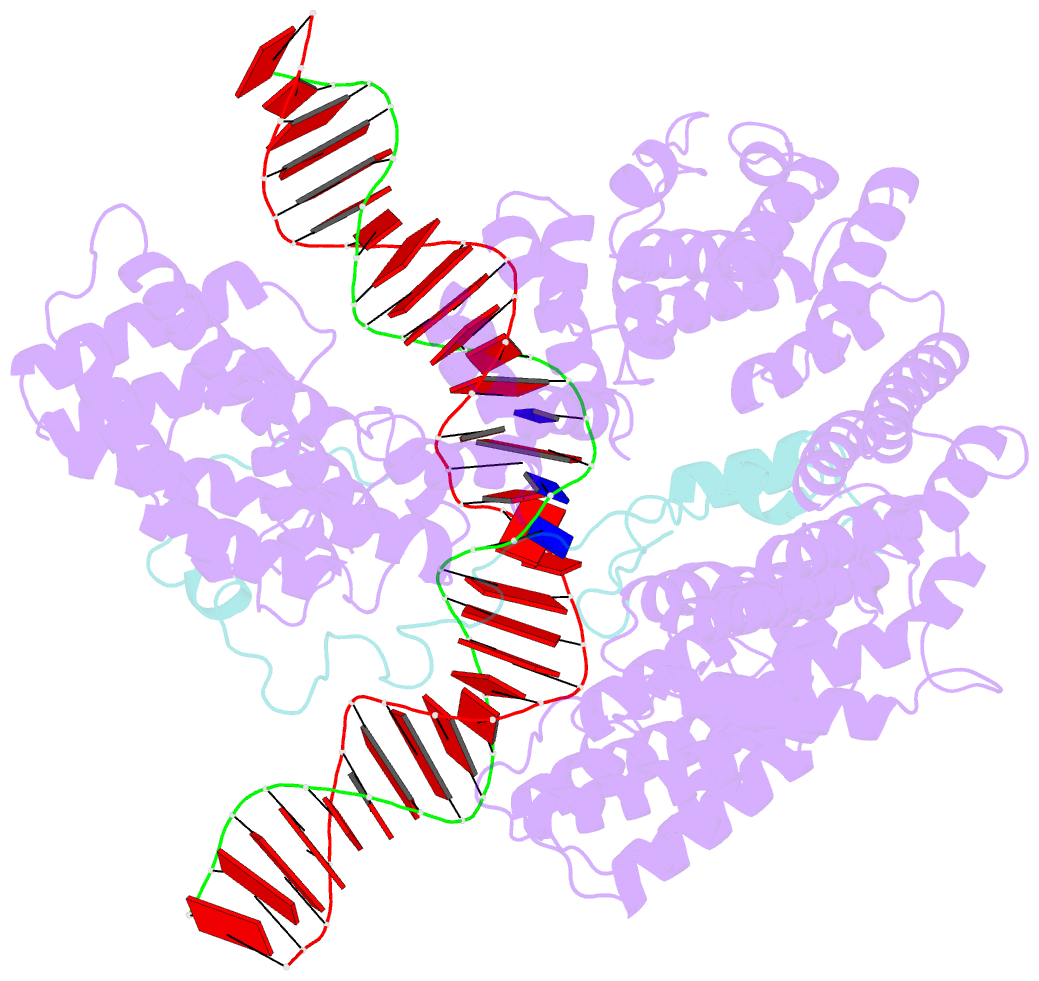
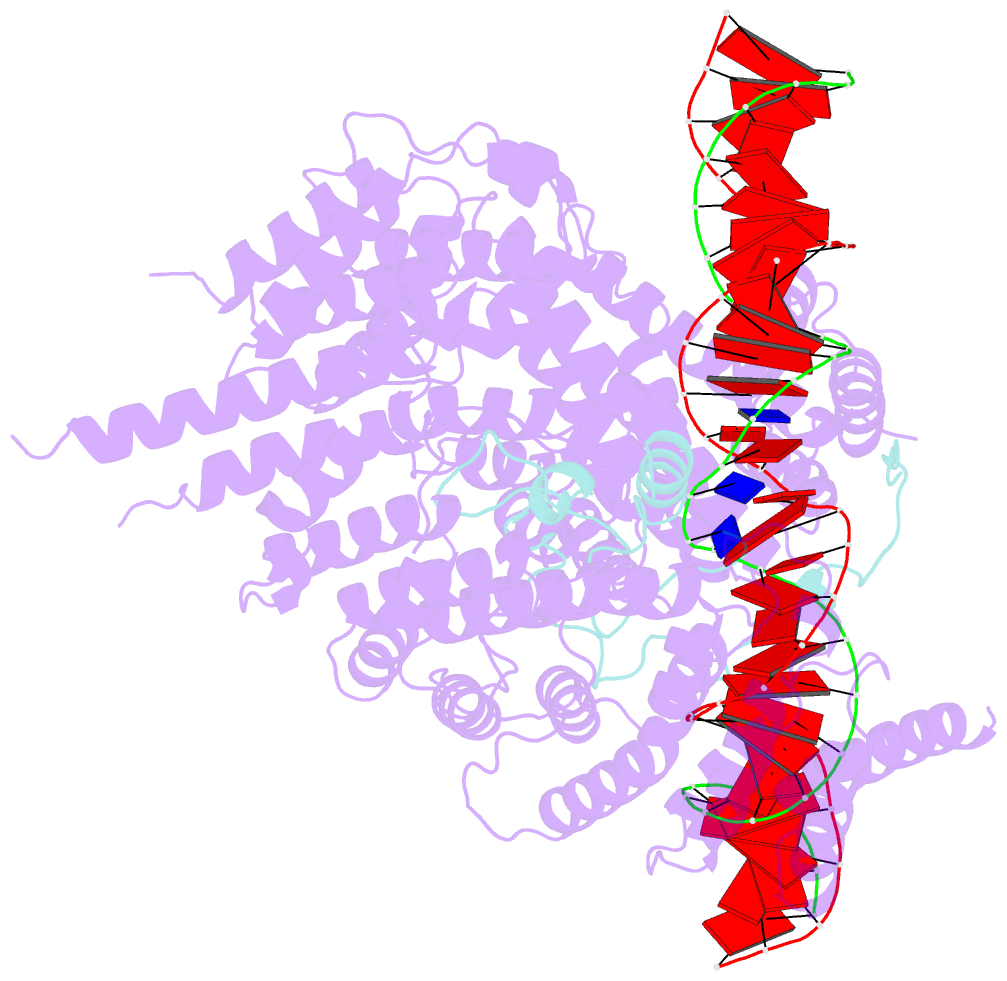
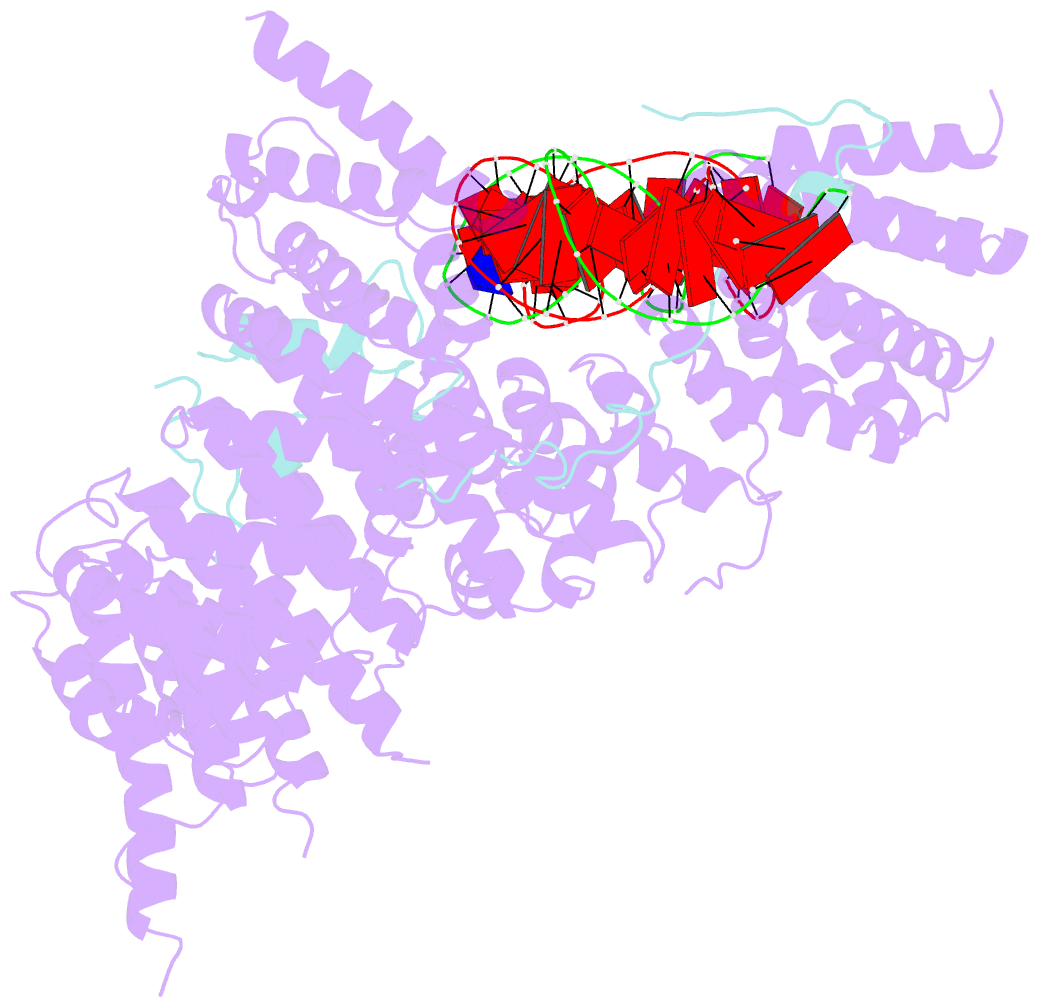
- S.c. condensin peripheral ycg1 subcomplex bound to DNA
- Reference
- Shaltiel IA, Datta S, Lecomte L, Hassler M, Kschonsak M, Bravo S, Stober C, Ormanns J, Eustermann S, Haering CH (2022): "A hold-and-feed mechanism drives directional DNA loop extrusion by condensin." Science, 376, 1087-1094. doi: 10.1126/science.abm4012.
- Abstract
- Structural maintenance of chromosomes (SMC) protein complexes structure genomes by extruding DNA loops, but the molecular mechanism that underlies their activity has remained unknown. We show that the active condensin complex entraps the bases of a DNA loop transiently in two separate chambers. Single-molecule imaging and cryo-electron microscopy suggest a putative power-stroke movement at the first chamber that feeds DNA into the SMC-kleisin ring upon adenosine triphosphate binding, whereas the second chamber holds on upstream of the same DNA double helix. Unlocking the strict separation of "motor" and "anchor" chambers turns condensin from a one-sided into a bidirectional DNA loop extruder. We conclude that the orientation of two topologically bound DNA segments during the SMC reaction cycle determines the directionality of DNA loop extrusion.