Summary information and primary citation
- PDB-id
- 7r8h; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (2.54 Å)
- Summary
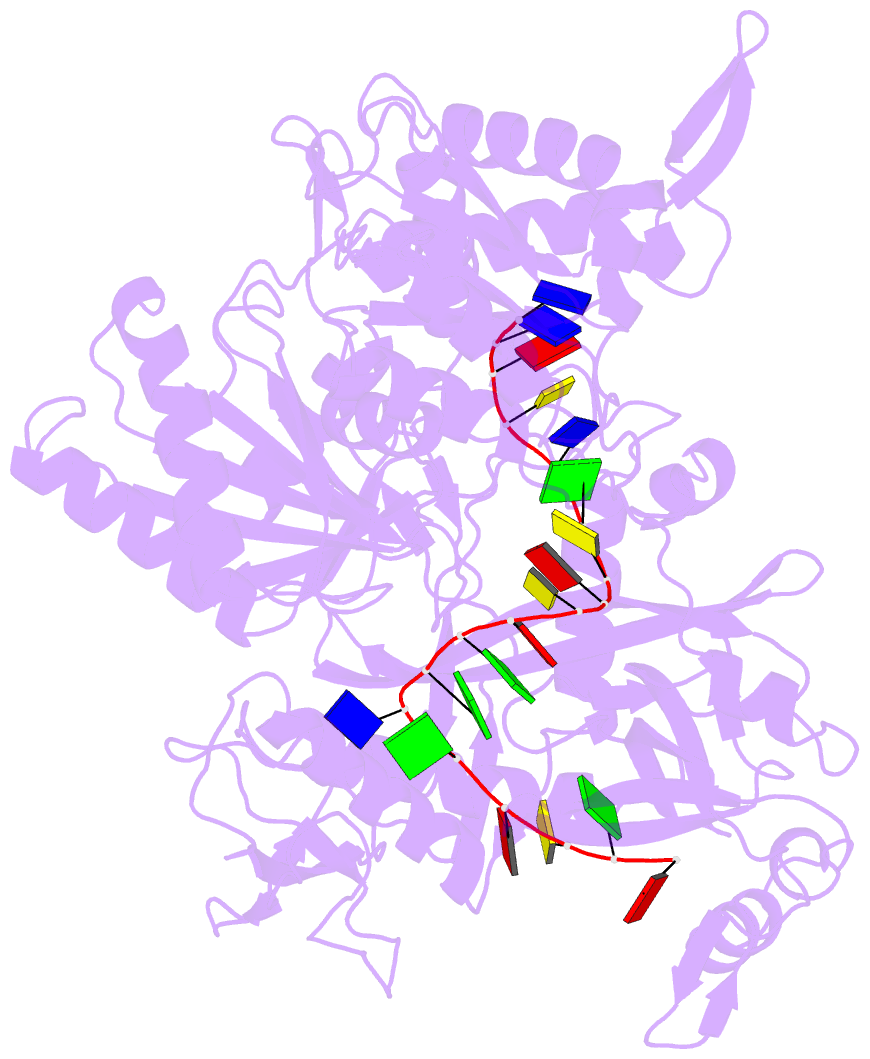
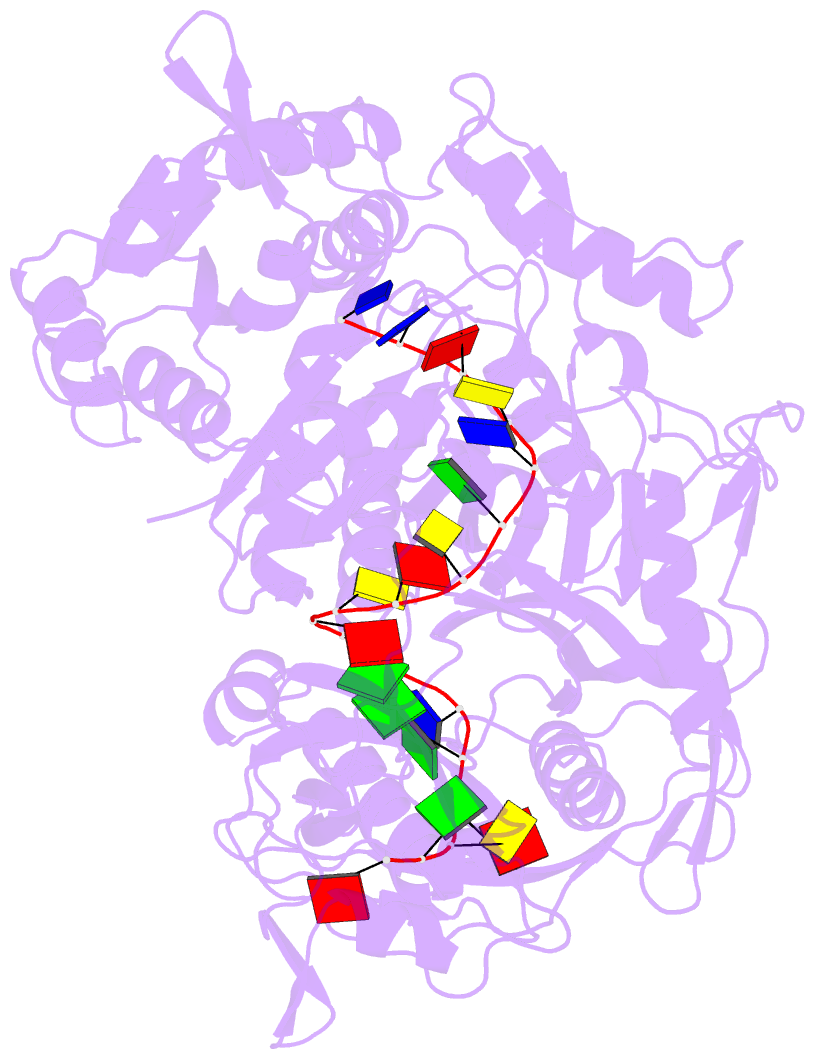
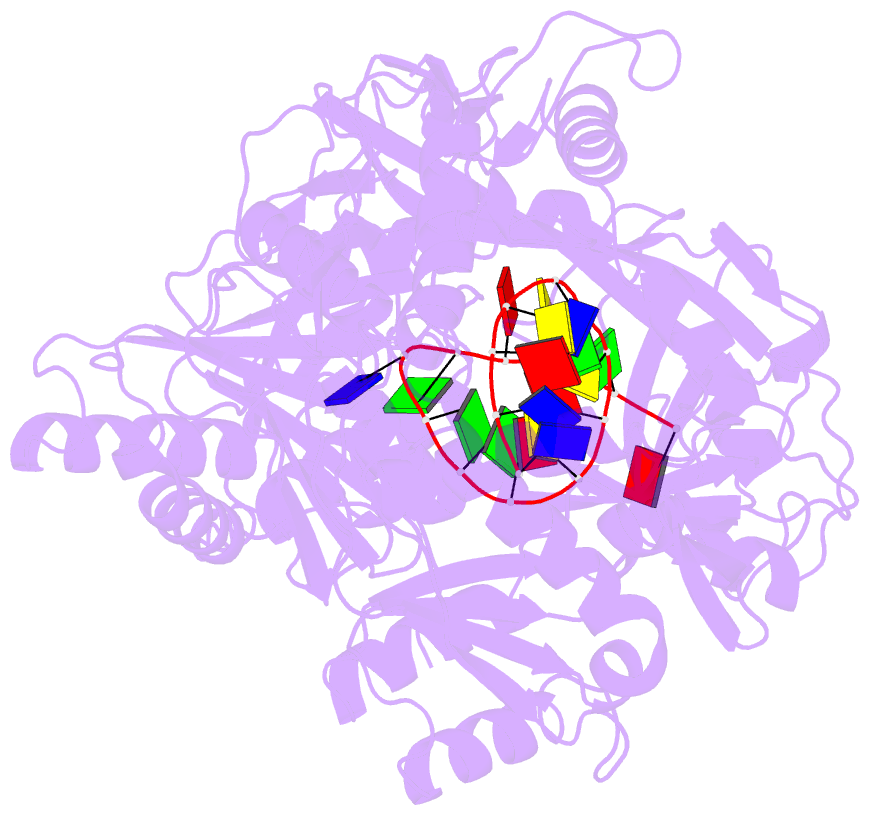
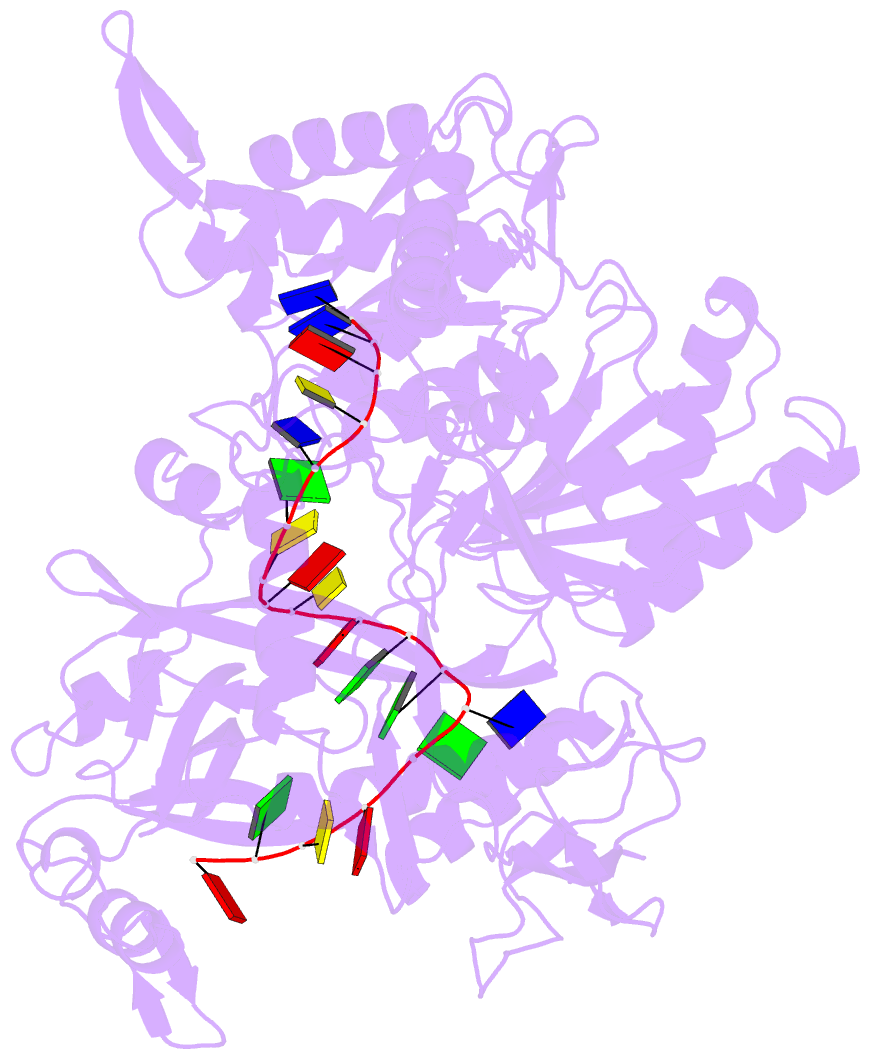
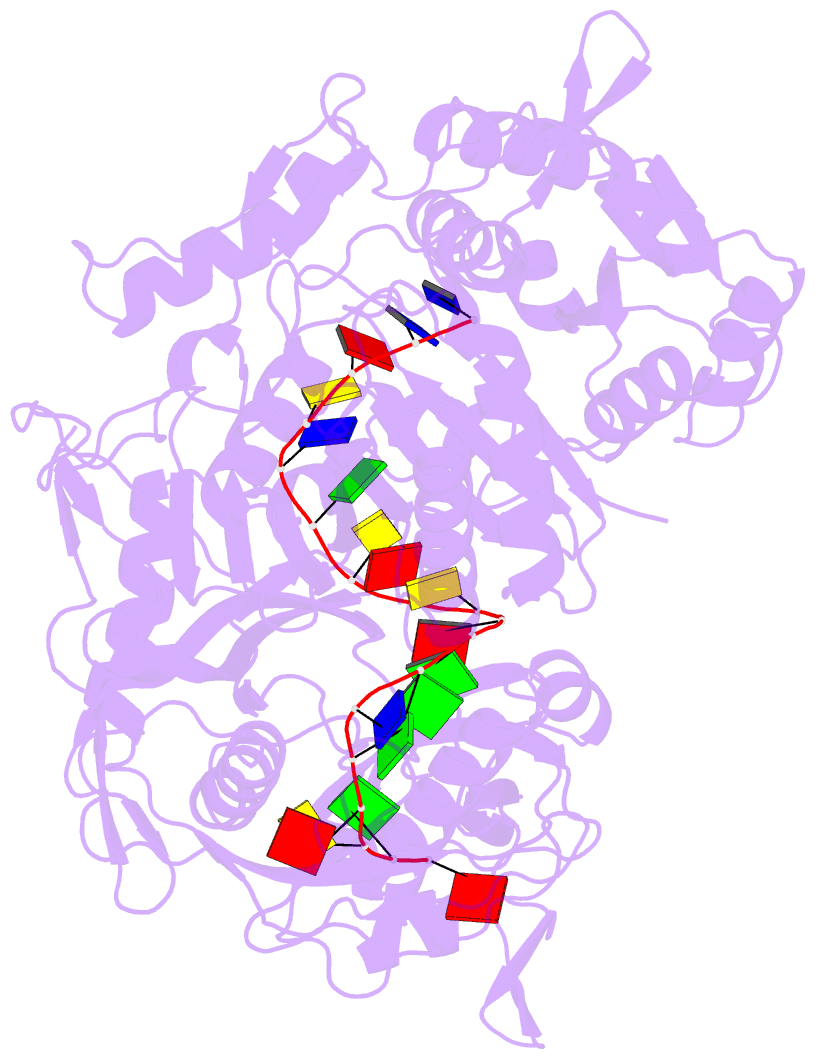
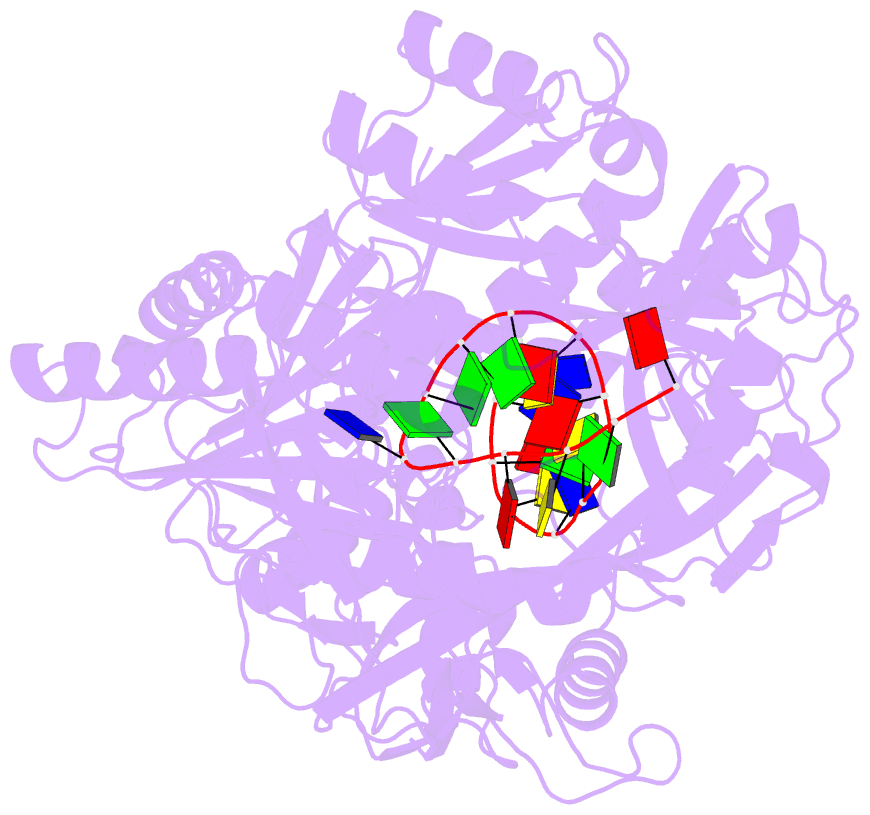
- Crystal structure of pseudooceanicola lipolyticus argonaute bound to 5' p guide DNA
- Reference
- Lisitskaya L, Shin Y, Agapov A, Olina A, Kropocheva E, Ryazansky S, Aravin AA, Esyunina D, Murakami KS, Kulbachinskiy A (2022): "Programmable RNA targeting by bacterial Argonaute nucleases with unconventional guide binding and cleavage specificity." Nat Commun, 13, 4624. doi: 10.1038/s41467-022-32079-5.
- Abstract
- Argonaute proteins are programmable nucleases that have defense and regulatory functions in both eukaryotes and prokaryotes. All known prokaryotic Argonautes (pAgos) characterized so far act on DNA targets. Here, we describe a new class of pAgos that uniquely use DNA guides to process RNA targets. The biochemical and structural analysis of Pseudooceanicola lipolyticus pAgo (PliAgo) reveals an unusual organization of the guide binding pocket that does not rely on divalent cations and the canonical set of contacts for 5'-end interactions. Unconventional interactions of PliAgo with the 5'-phosphate of guide DNA define its new position within pAgo and shift the site of target RNA cleavage in comparison with known Argonautes. The specificity for RNA over DNA is defined by ribonucleotide residues at the cleavage site. The analysed pAgos sense mismatches and modifications in the RNA target. The results broaden our understanding of prokaryotic defense systems and extend the spectrum of programmable nucleases with potential use in RNA technology.