Summary information and primary citation
- PDB-id
- 7r9g; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- X-ray (2.4 Å)
- Summary
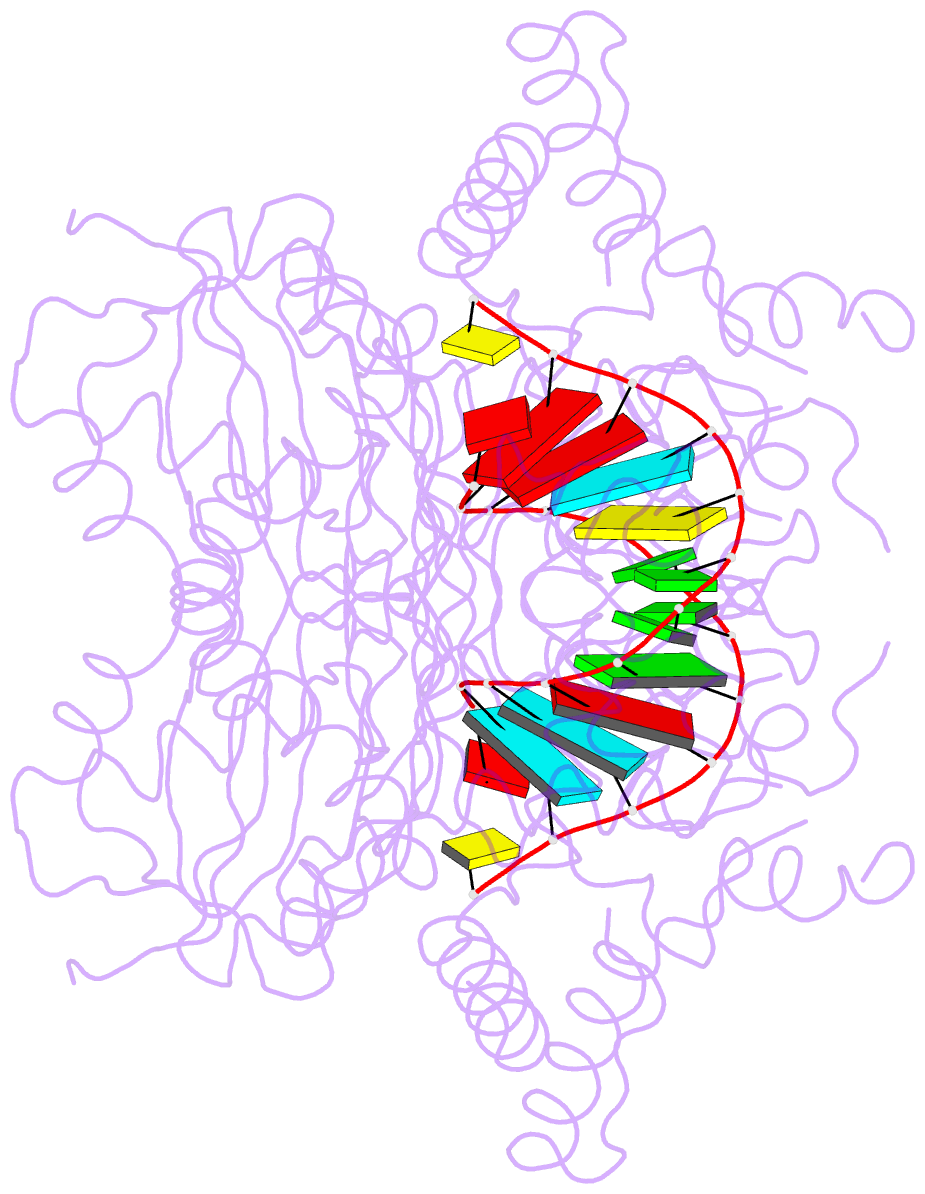
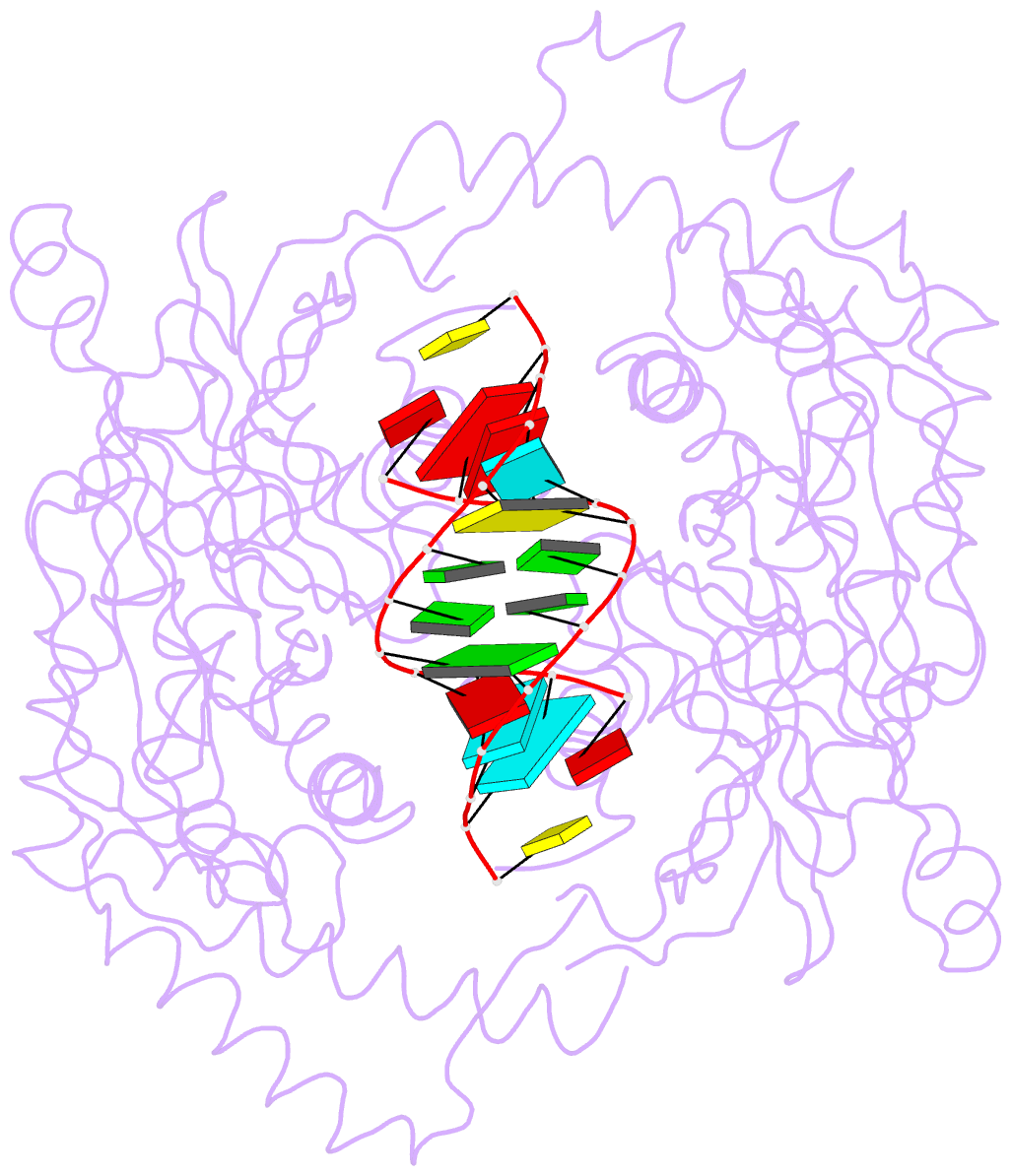
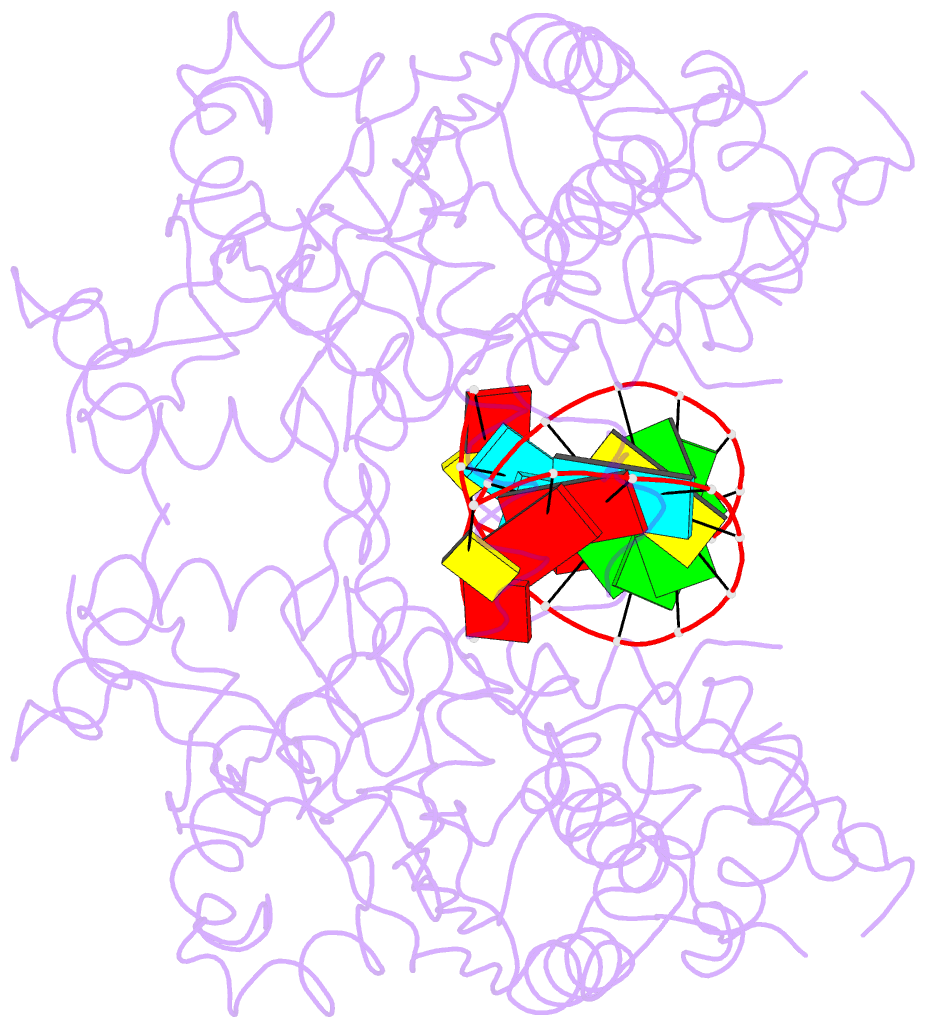
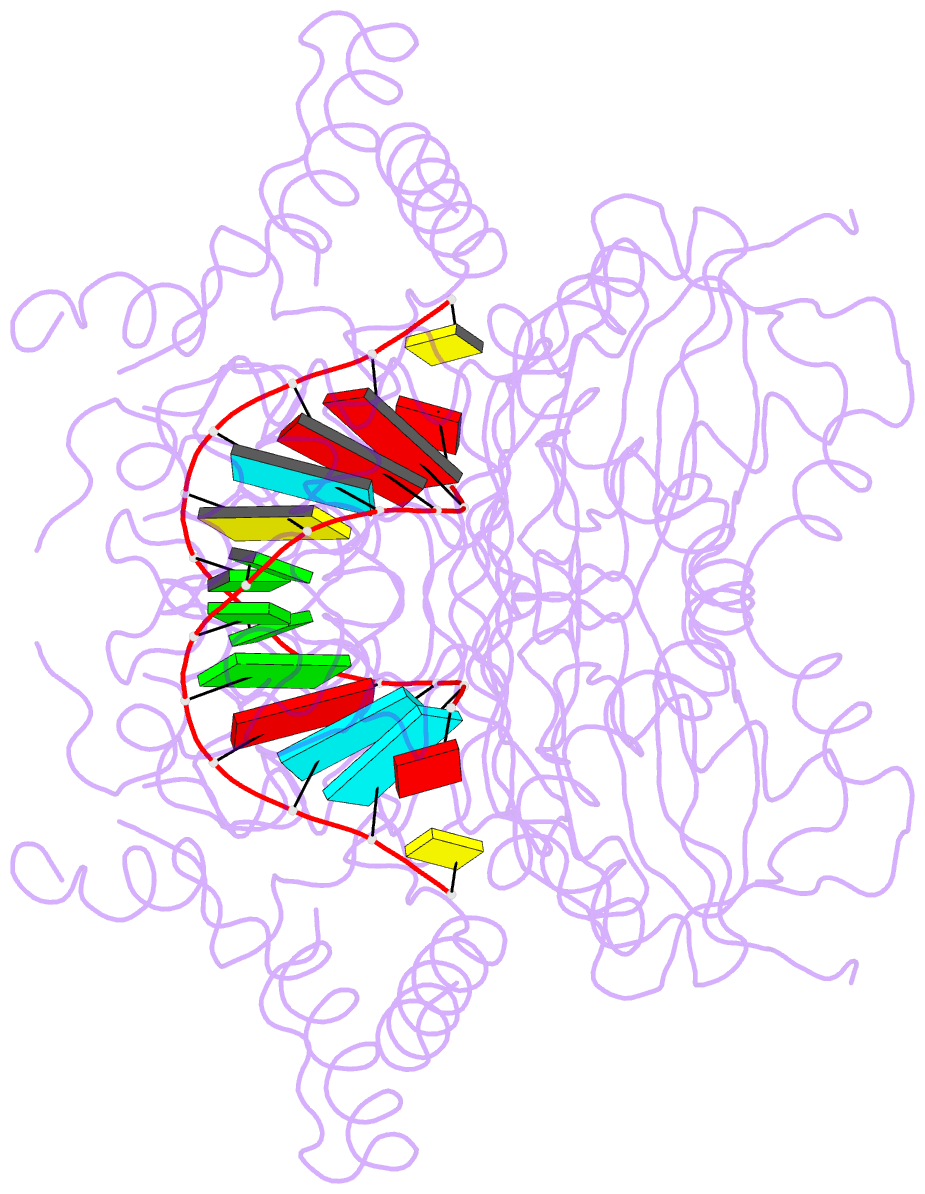
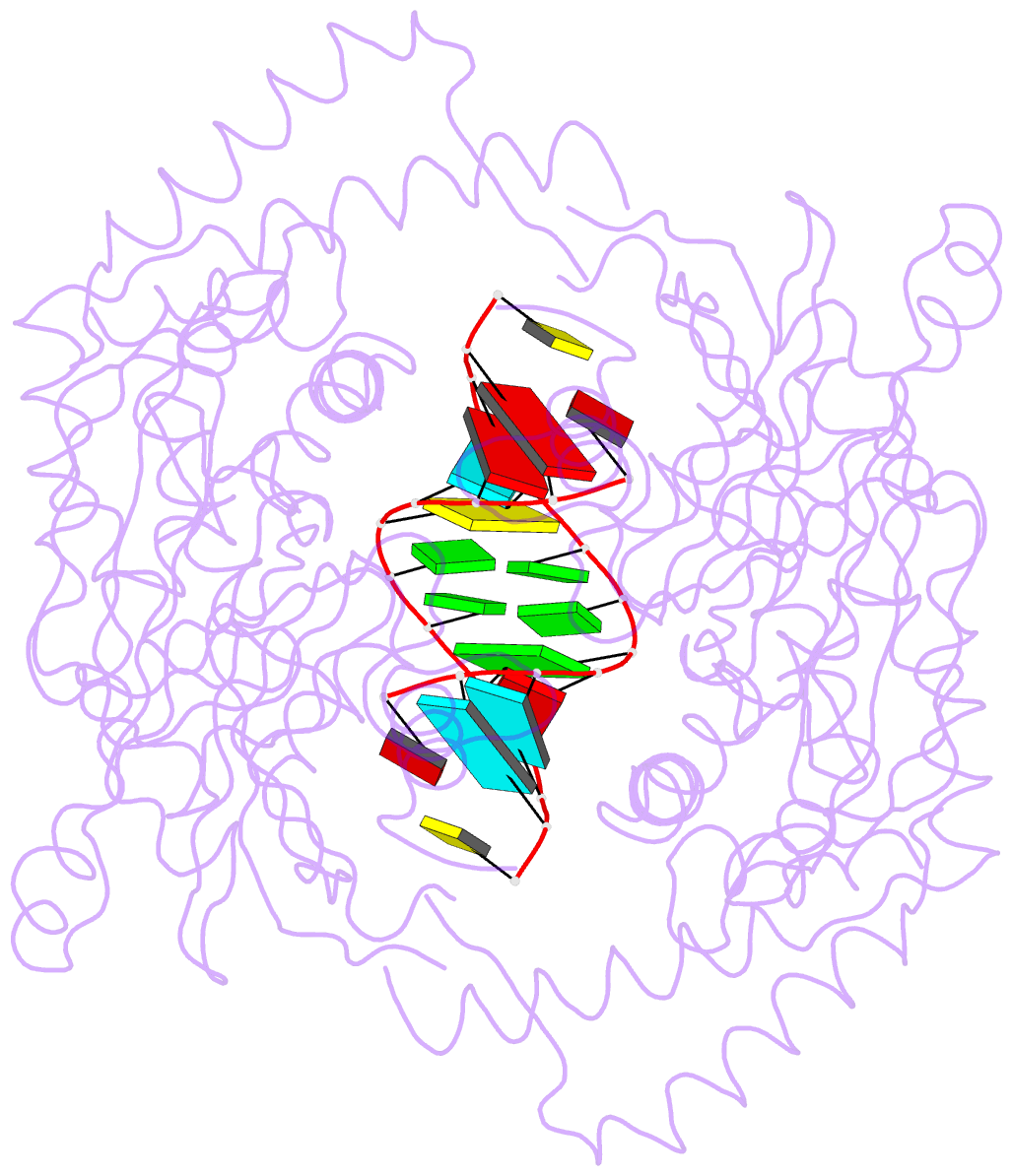
- Catalytically inactive yeast pseudouridine synthase, pus1, bound to RNA
- Reference
- Grunberg S, Doyle LA, Wolf EJ, Dai N, Correa Jr IR, Yigit E, Stoddard BL (2023): "The structural basis of mRNA recognition and binding by yeast pseudouridine synthase PUS1." Plos One, 18, e0291267. doi: 10.1371/journal.pone.0291267.
- Abstract
- The chemical modification of RNA bases represents a ubiquitous activity that spans all domains of life. Pseudouridylation is the most common RNA modification and is observed within tRNA, rRNA, ncRNA and mRNAs. Pseudouridine synthase or 'PUS' enzymes include those that rely on guide RNA molecules and others that function as 'stand-alone' enzymes. Among the latter, several have been shown to modify mRNA transcripts. Although recent studies have defined the structural requirements for RNA to act as a PUS target, the mechanisms by which PUS1 recognizes these target sequences in mRNA are not well understood. Here we describe the crystal structure of yeast PUS1 bound to an RNA target that we identified as being a hot spot for PUS1-interaction within a model mRNA at 2.4 Å resolution. The enzyme recognizes and binds both strands in a helical RNA duplex, and thus guides the RNA containing the target uridine to the active site for subsequent modification of the transcript. The study also allows us to show the divergence of related PUS1 enzymes and their corresponding RNA target specificities, and to speculate on the basis by which PUS1 binds and modifies mRNA or tRNA substrates.