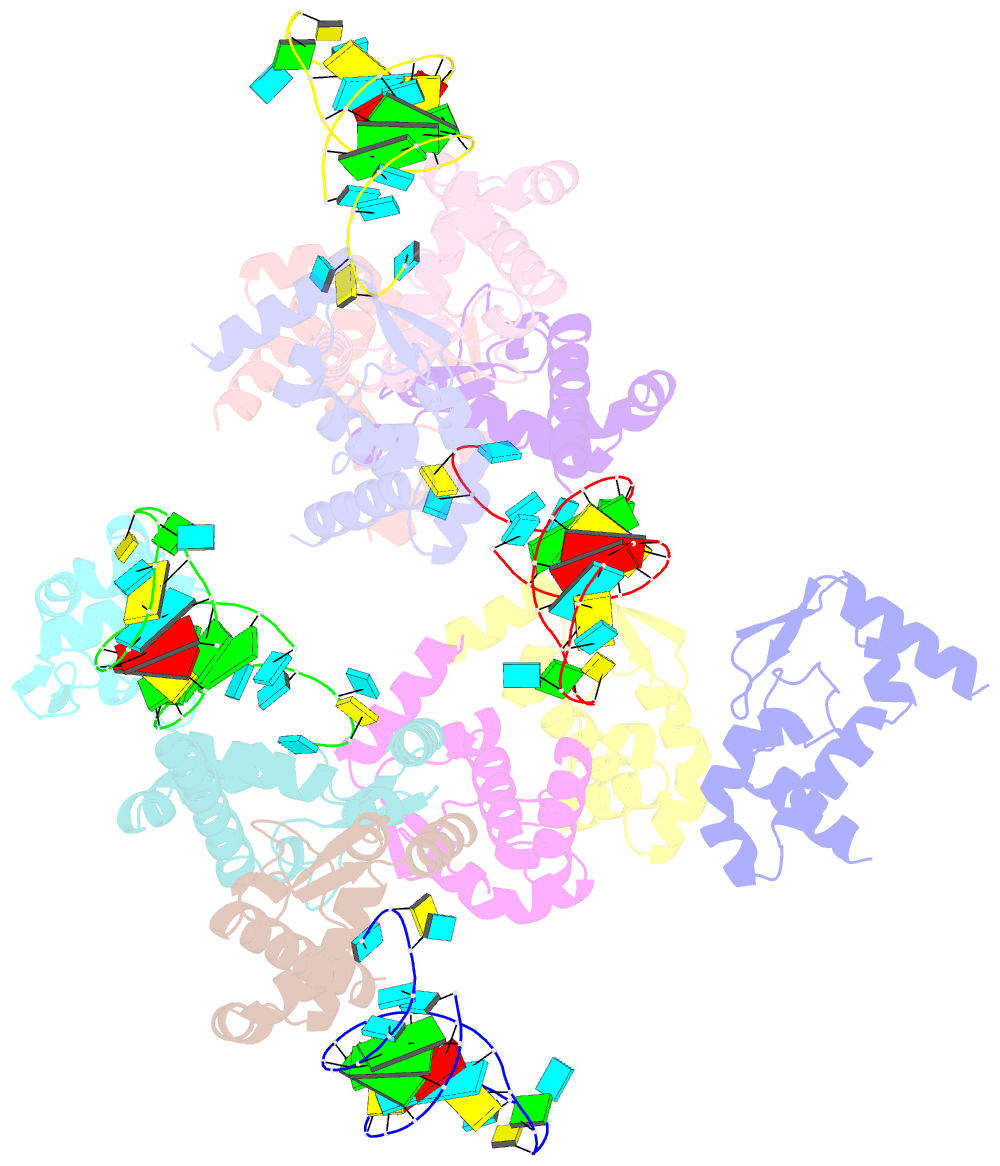
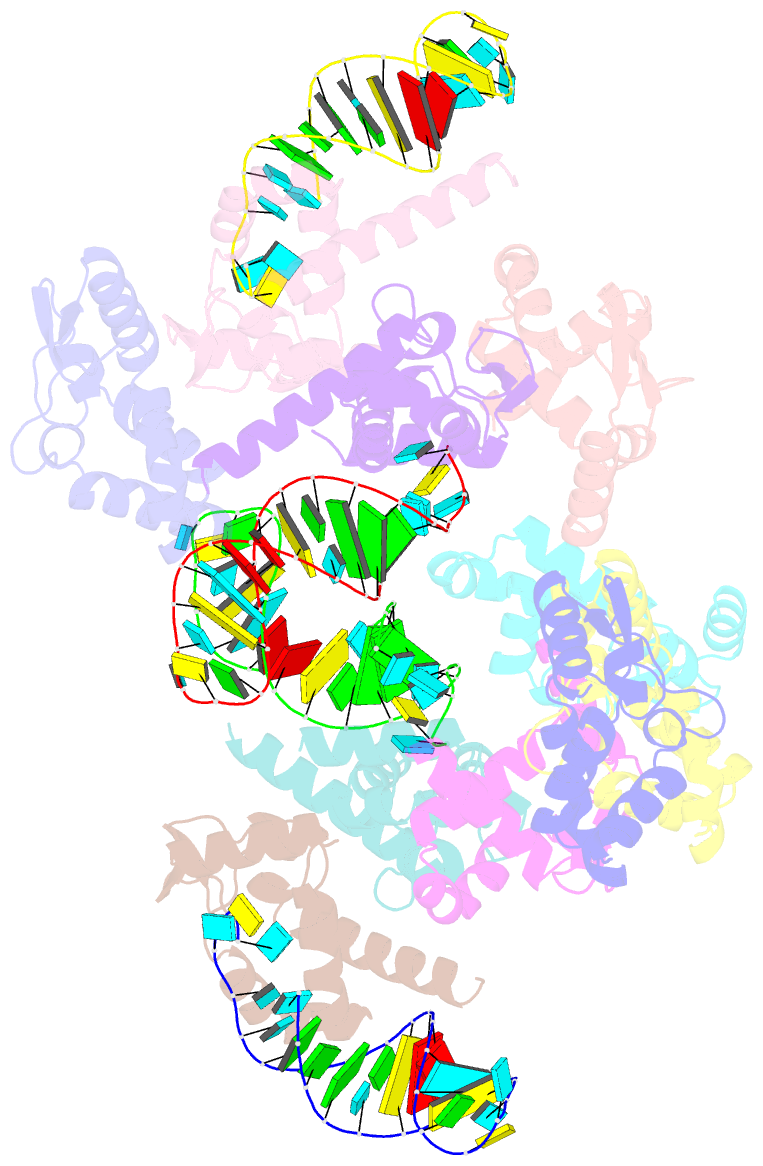
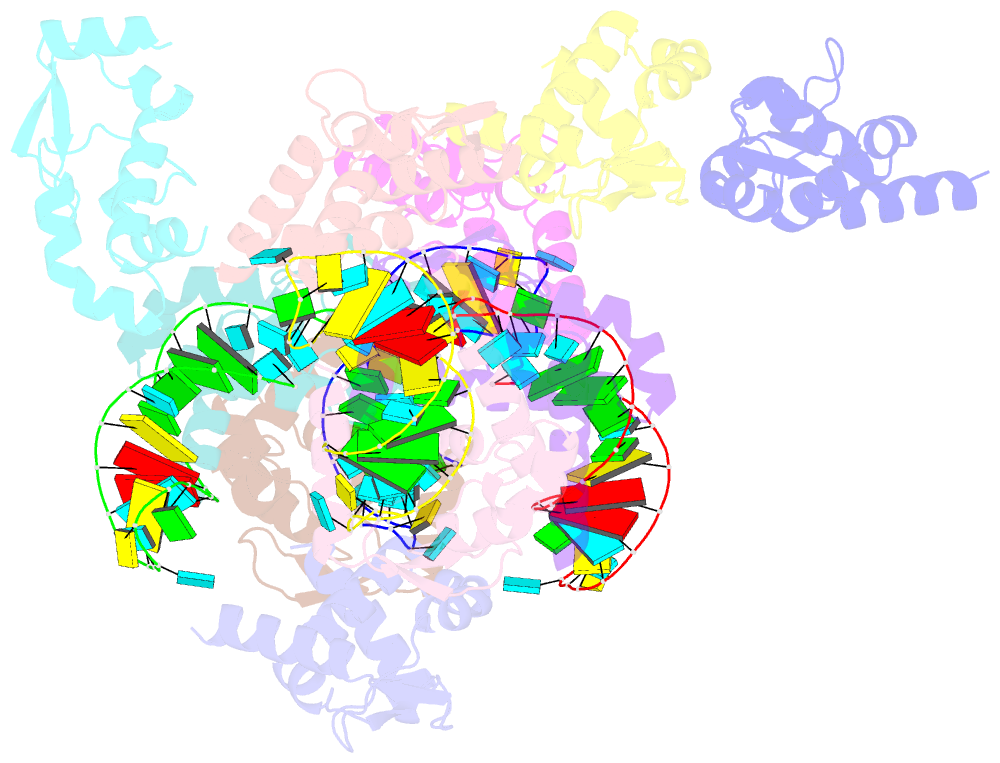
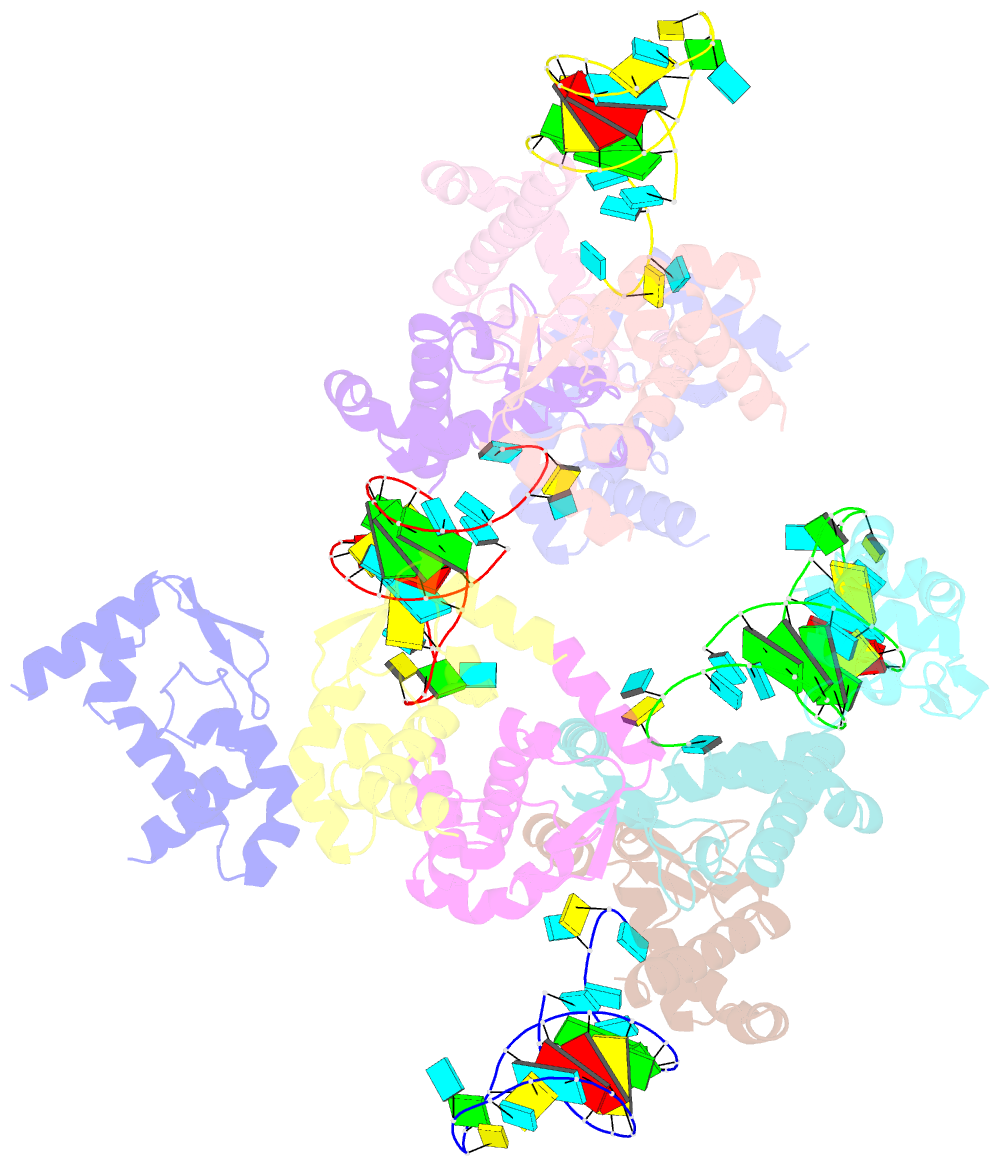
Summary information and primary citation
- PDB-id
- 7rgu; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- X-ray (3.2 Å)
- Summary
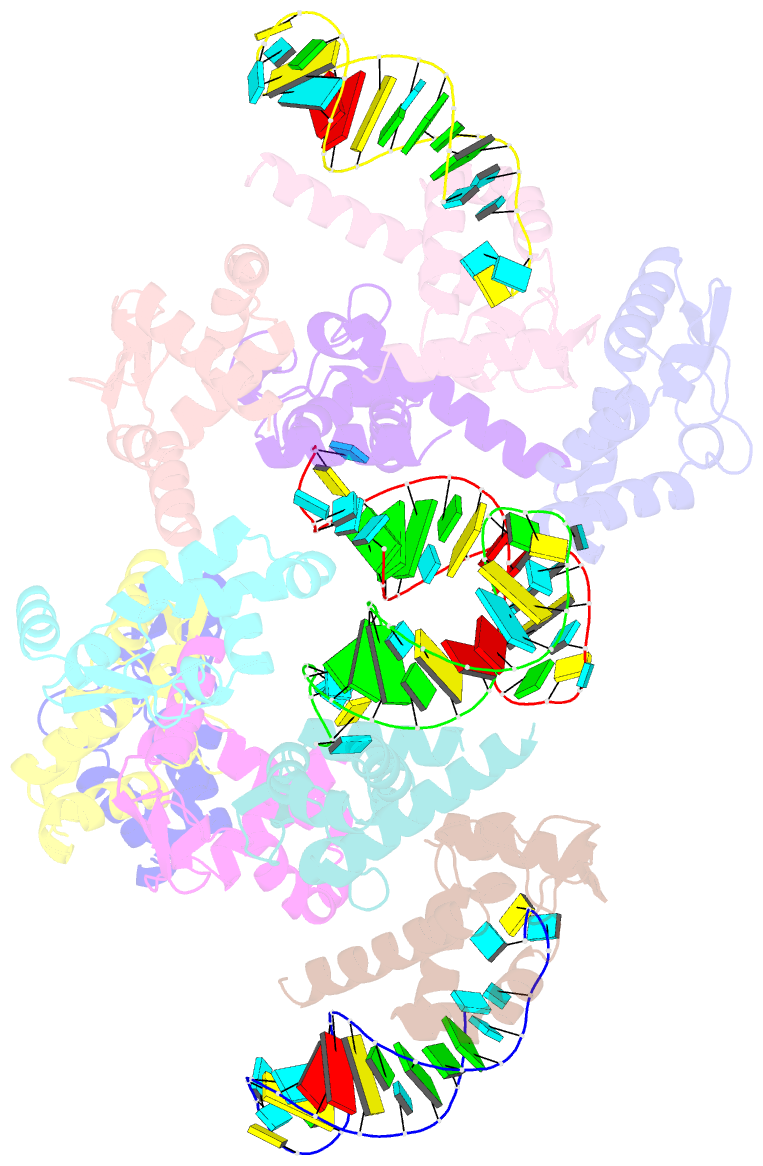
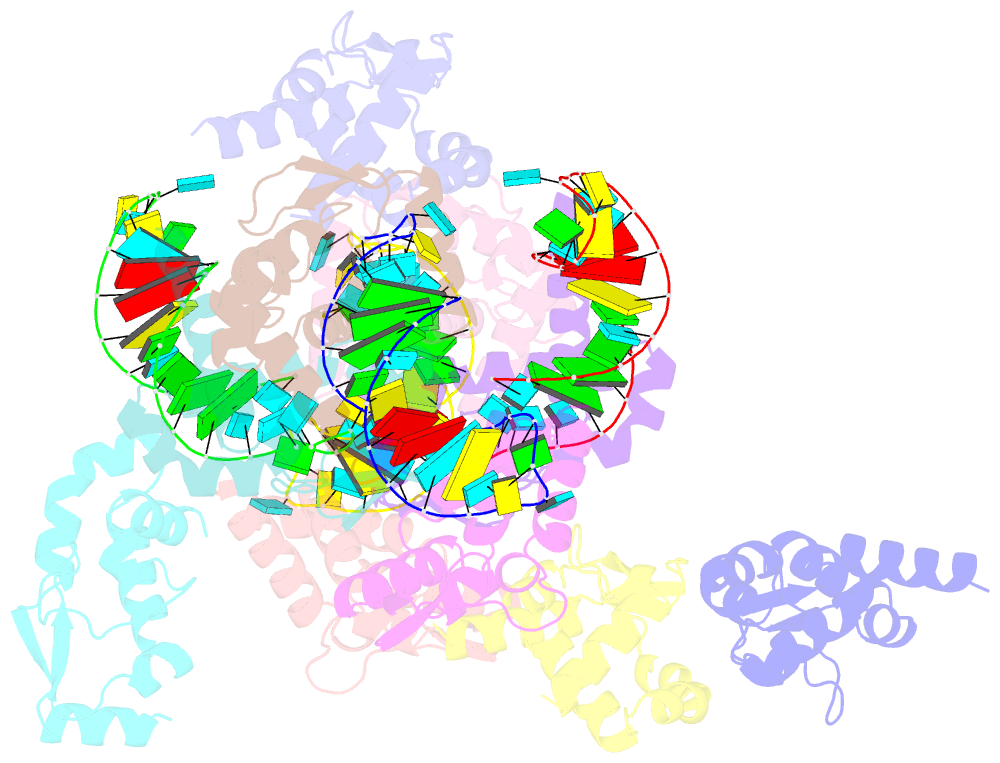
- The crystal structure of rocc bound to a transcriptional terminator
- Reference
- Kim HJ, Black M, Edwards RA, Peillard-Fiorente F, Panigrahi R, Klingler D, Eidelpes R, Zeindl R, Peng S, Su J, Omar AR, MacMillan AM, Kreutz C, Tollinger M, Charpentier X, Attaiech L, Glover JNM (2022): "Structural basis for recognition of transcriptional terminator structures by ProQ/FinO domain RNA chaperones." Nat Commun, 13, 7076. doi: 10.1038/s41467-022-34875-5.
- Abstract
- The ProQ/FinO family of RNA binding proteins mediate sRNA-directed gene regulation throughout gram-negative bacteria. Here, we investigate the structural basis for RNA recognition by ProQ/FinO proteins, through the crystal structure of the ProQ/FinO domain of the Legionella pneumophila DNA uptake regulator, RocC, bound to the transcriptional terminator of its primary partner, the sRNA RocR. The structure reveals specific recognition of the 3' nucleotide of the terminator by a conserved pocket involving a β-turn-α-helix motif, while the hairpin portion of the terminator is recognized by a conserved α-helical N-cap motif. Structure-guided mutagenesis reveals key RNA contact residues that are critical for RocC/RocR to repress the uptake of environmental DNA in L. pneumophila. Structural analysis and RNA binding studies reveal that other ProQ/FinO domains also recognize related transcriptional terminators with different specificities for the length of the 3' ssRNA tail.