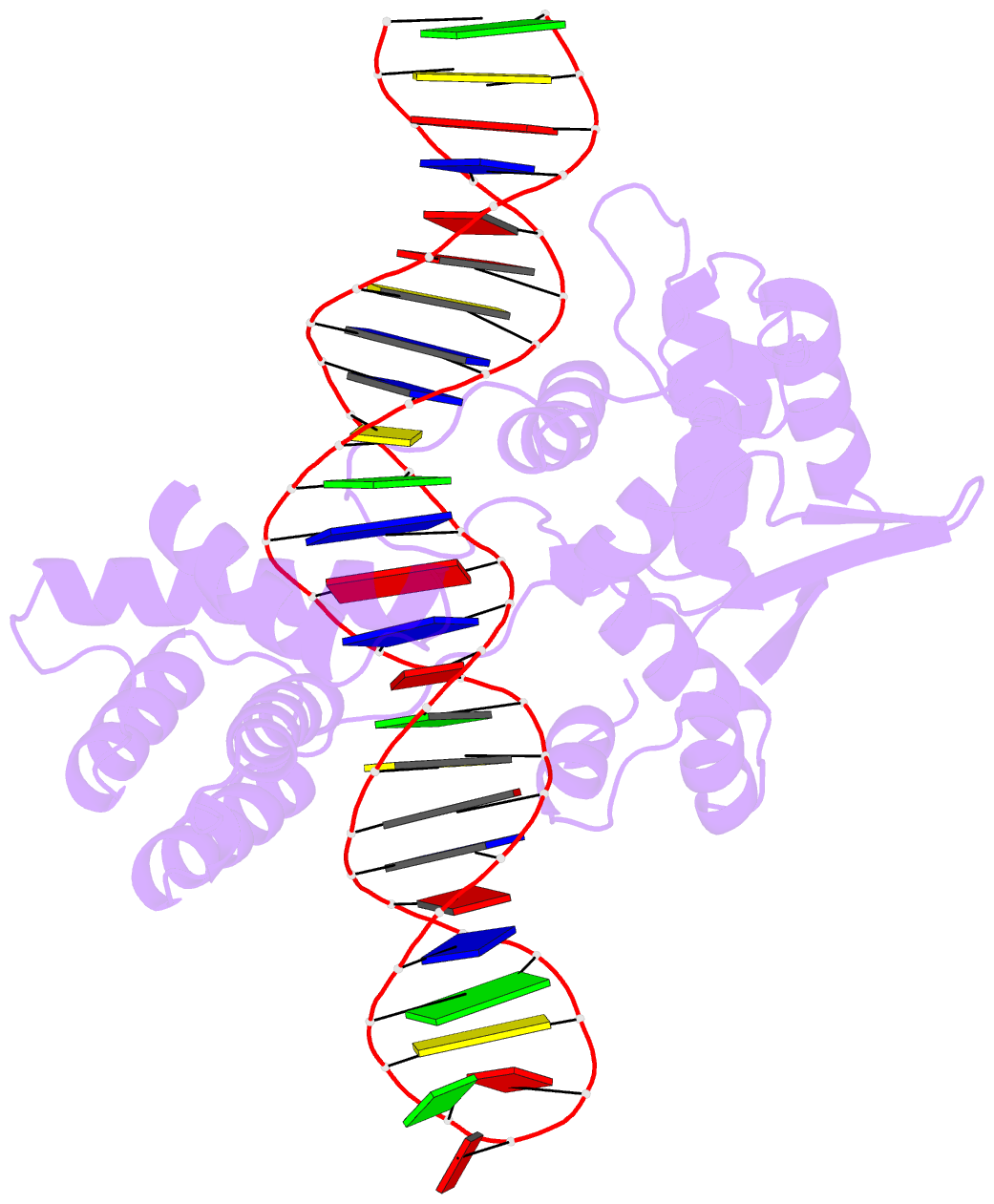
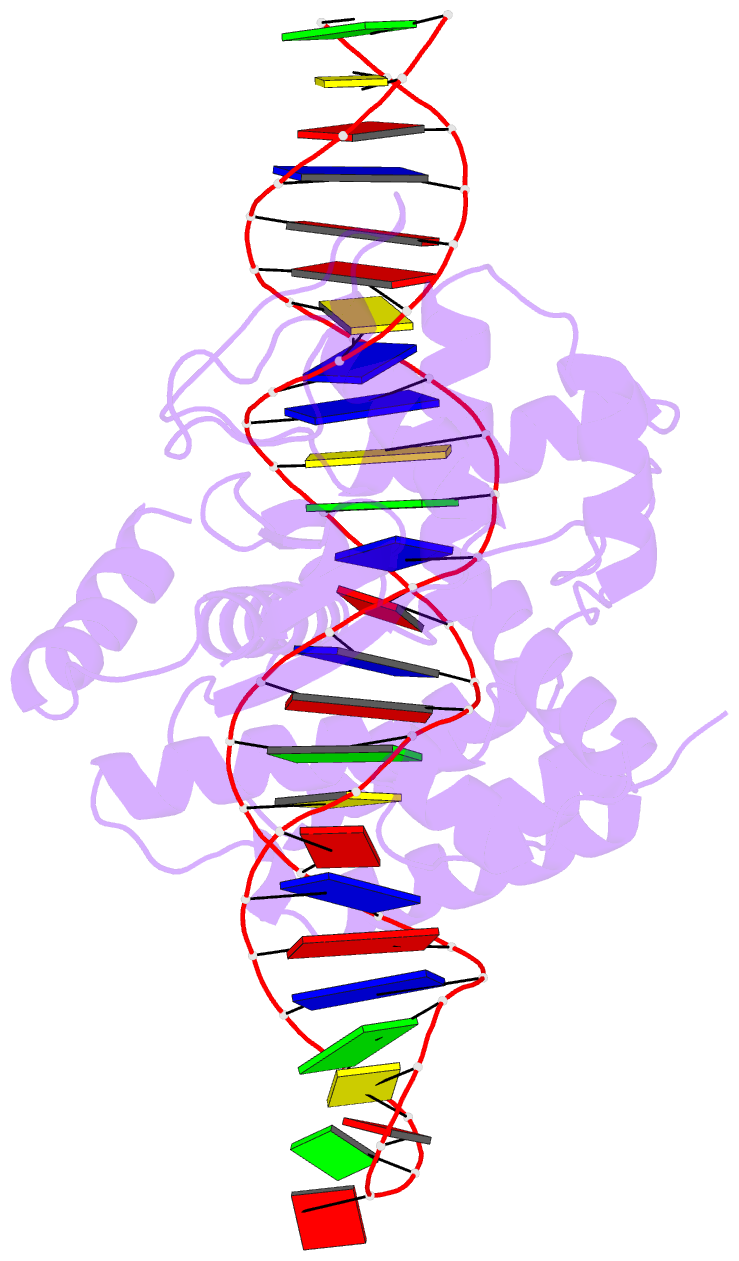
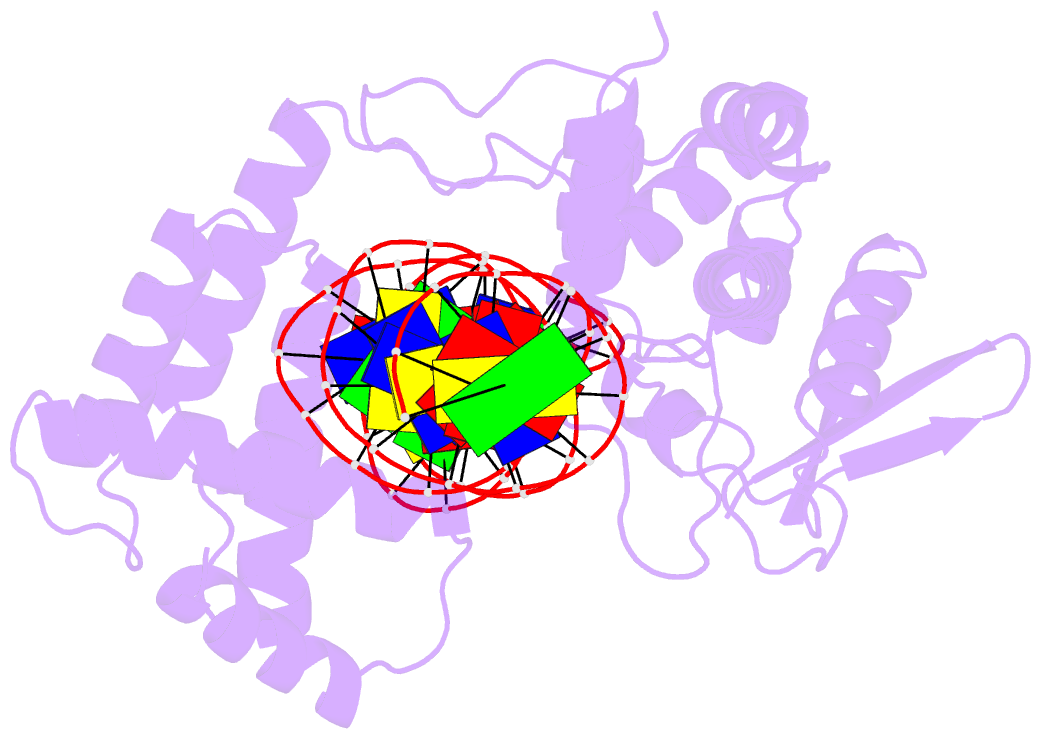
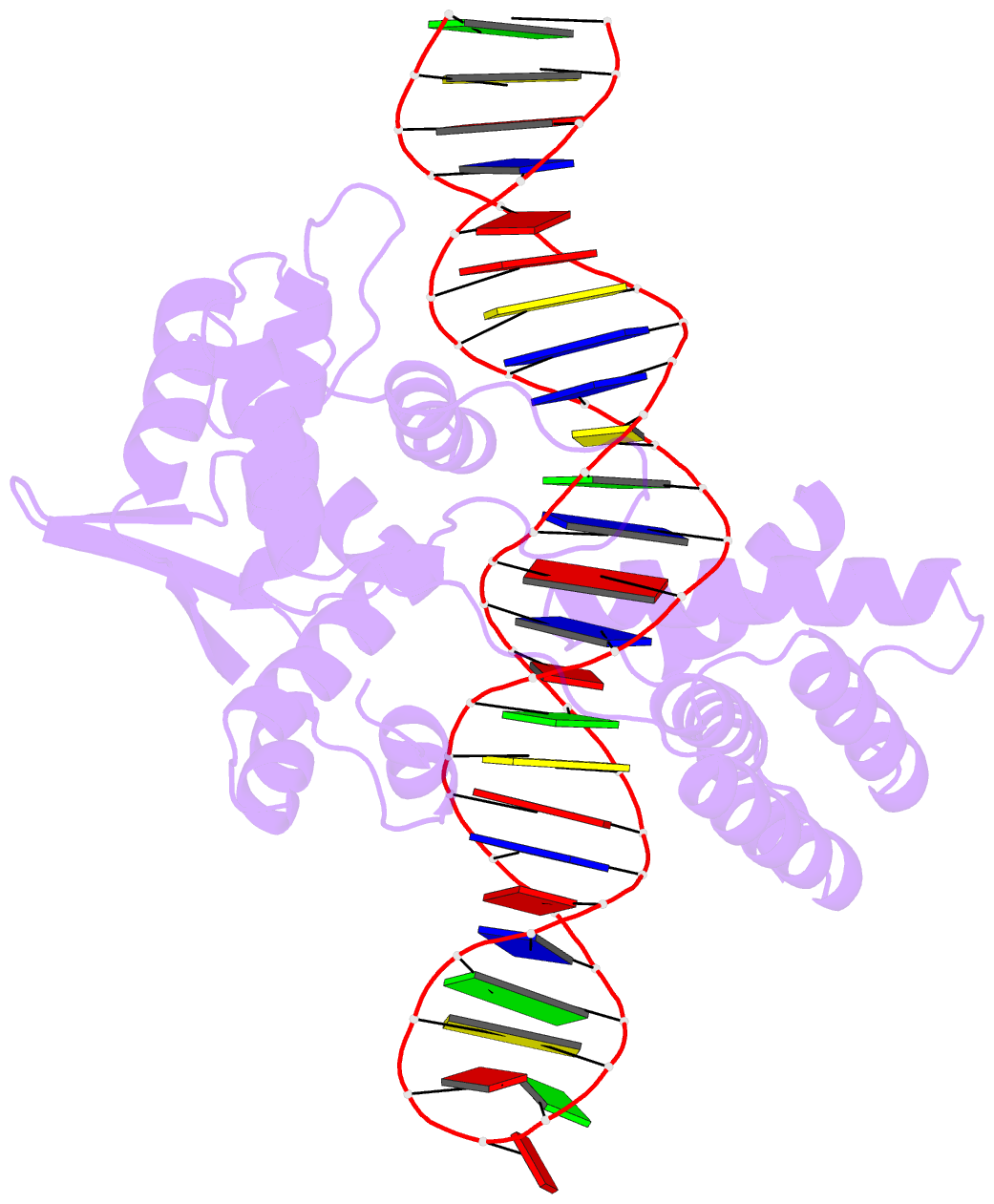
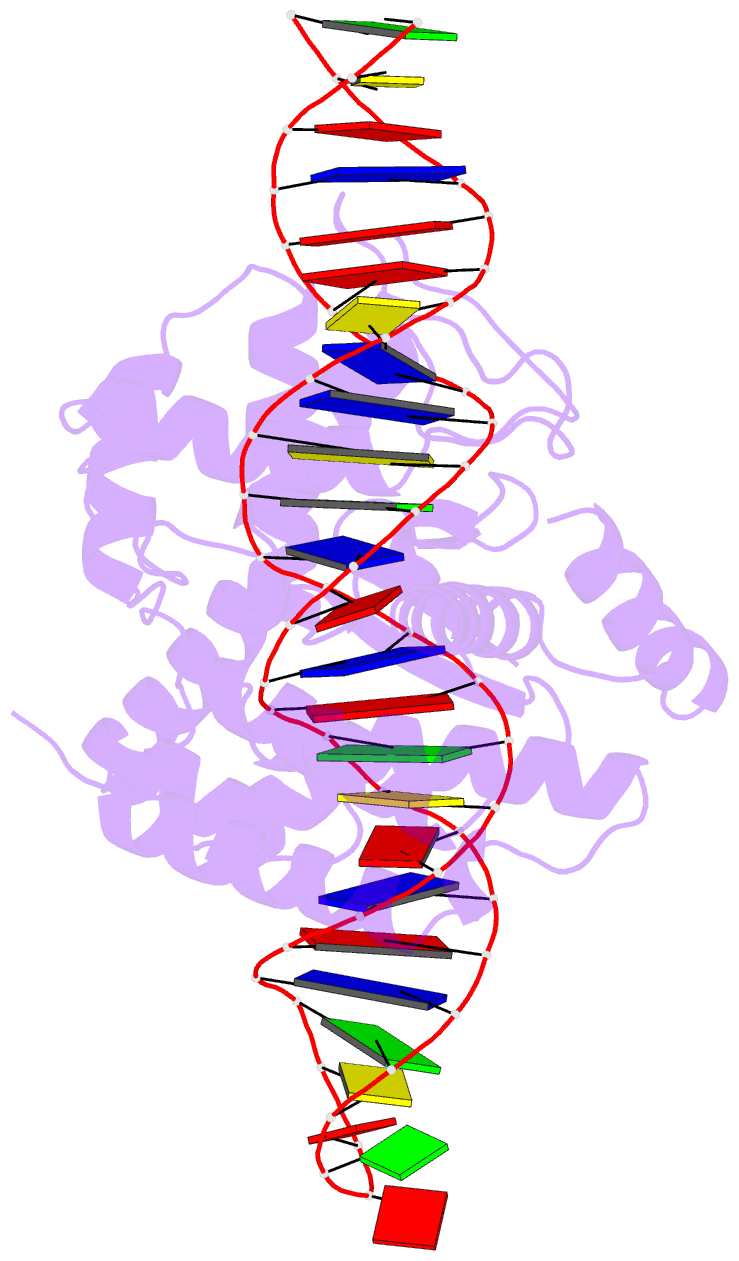
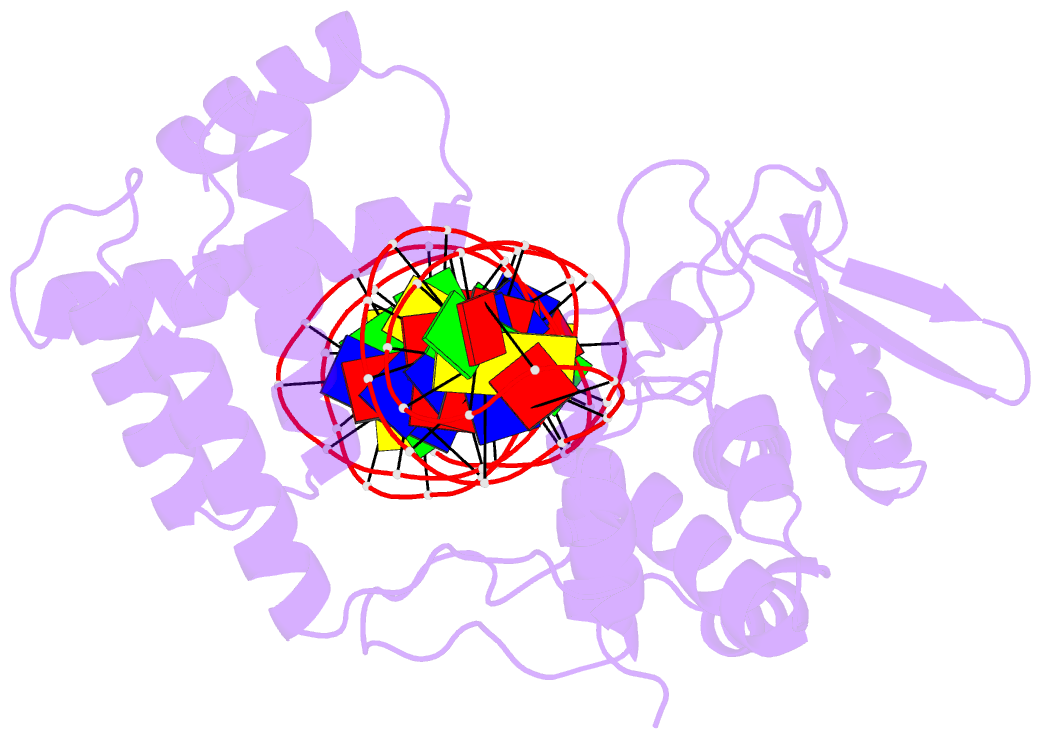
Summary information and primary citation
- PDB-id
- 7rhy; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- recombination-DNA
- Method
- cryo-EM (3.91 Å)
- Summary
- Cre recombinase mutant (d33a-a36v-r192a) in complex with loxa DNA hairpin
- Reference
- Stachowski K, Norris AS, Potter D, Wysocki VH, Foster MP (2022): "Mechanisms of Cre recombinase synaptic complex assembly and activation illuminated by Cryo-EM." Nucleic Acids Res., 50, 1753-1769. doi: 10.1093/nar/gkac032.
- Abstract
- Cre recombinase selectively recognizes DNA and prevents non-specific DNA cleavage through an orchestrated series of assembly intermediates. Cre recombines two loxP DNA sequences featuring a pair of palindromic recombinase binding elements and an asymmetric spacer region, by assembly of a tetrameric synaptic complex, cleavage of an opposing pair of strands, and formation of a Holliday junction intermediate. We used Cre and loxP variants to isolate the monomeric Cre-loxP (54 kDa), dimeric Cre2-loxP (110 kDa), and tetrameric Cre4-loxP2 assembly intermediates, and determined their structures using cryo-EM to resolutions of 3.9, 4.5 and 3.2 Å, respectively. Progressive and asymmetric bending of the spacer region along the assembly pathway enables formation of increasingly intimate interfaces between Cre protomers and illuminates the structural bases of biased loxP strand cleavage order and half-the-sites activity. Application of 3D variability analysis to the tetramer data reveals constrained conformational sampling along the pathway between protomer activation and Holliday junction isomerization. These findings underscore the importance of protein and DNA flexibility in Cre-mediated site selection, controlled activation of alternating protomers, the basis for biased strand cleavage order, and recombination efficiency. Such considerations may advance development of site-specific recombinases for use in gene editing applications.