Summary information and primary citation
- PDB-id
- 7rzz; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein
- Method
- X-ray (2.39 Å)
- Summary
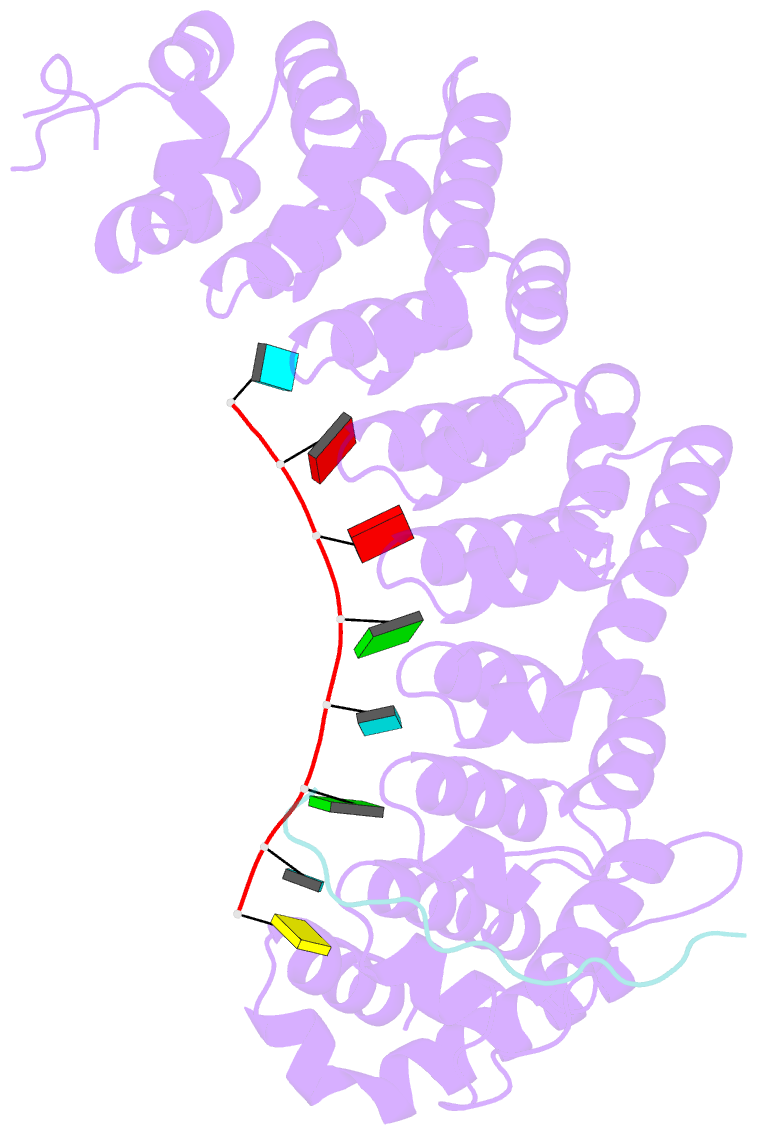
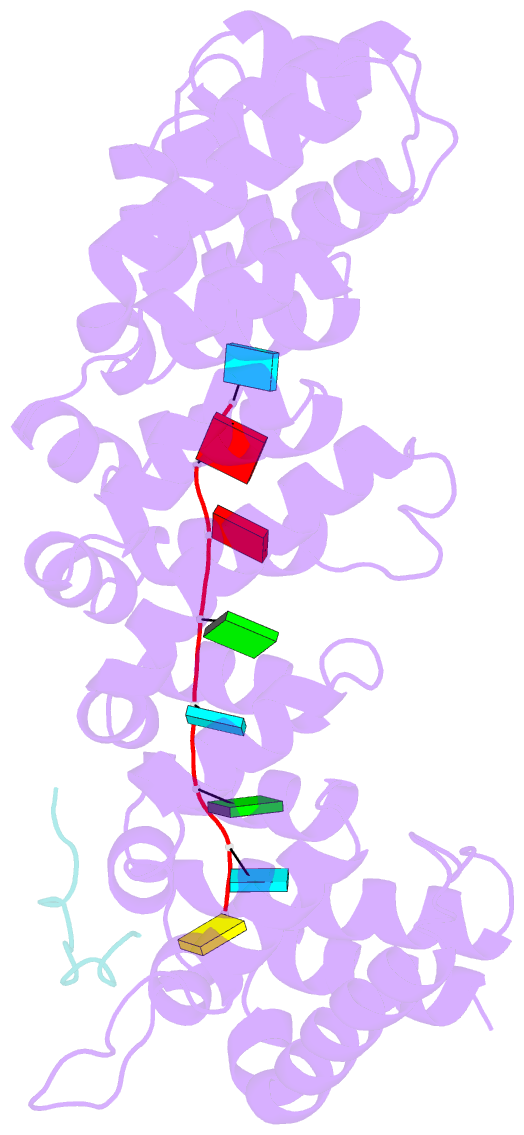
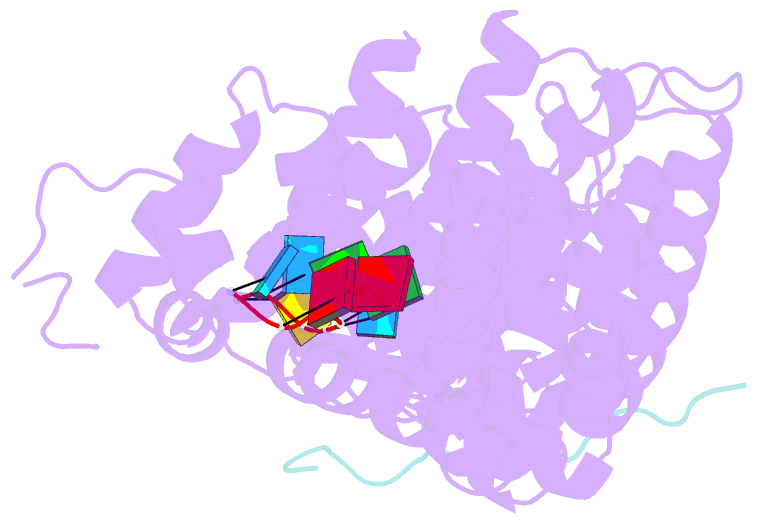
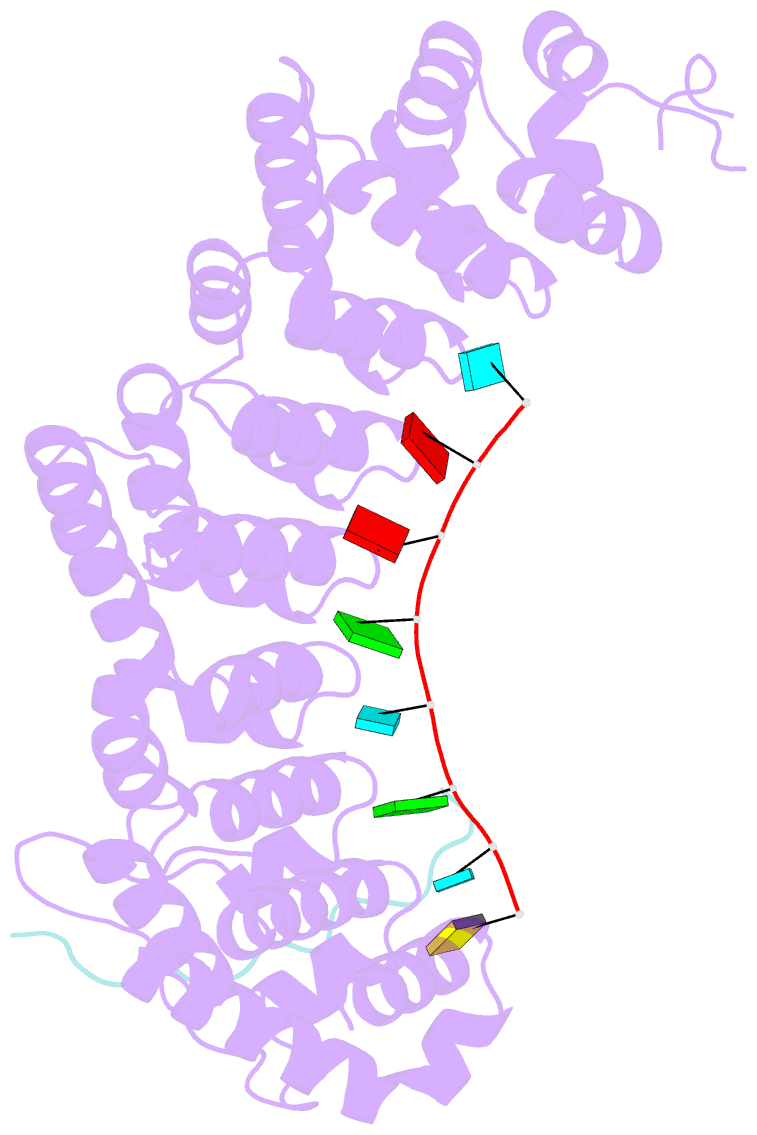
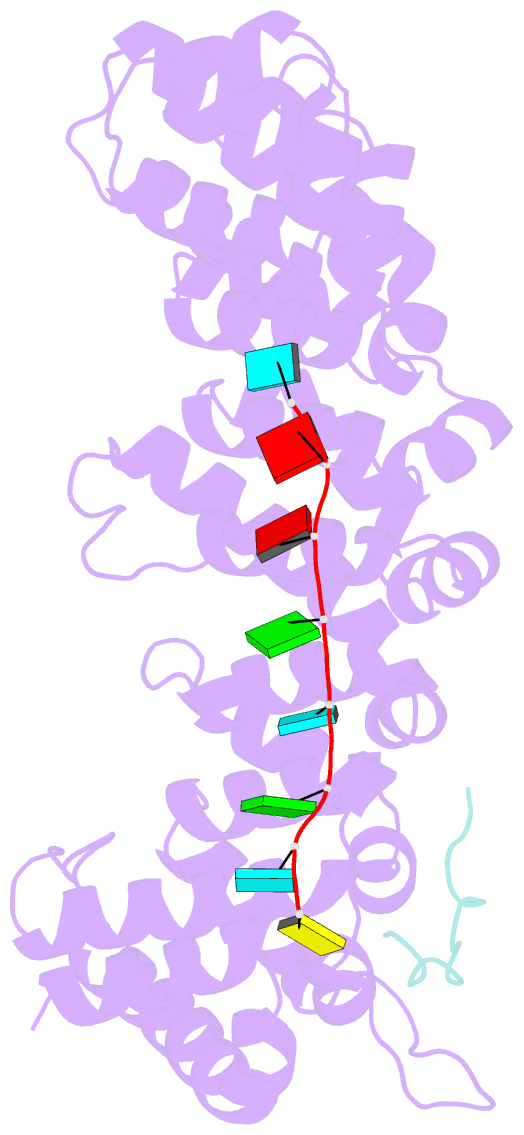
- Crystal structure of fbf-2 in complex with lst-1 site a peptide and compact fbe RNA
- Reference
- Qiu C, Wine RN, Campbell ZT, Hall TMT (2022): "Bipartite interaction sites differentially modulate RNA-binding affinity of a protein complex essential for germline stem cell self-renewal." Nucleic Acids Res., 50, 536-548. doi: 10.1093/nar/gkab1220.
- Abstract
- In C. elegans, PUF proteins promote germline stem cell self-renewal. Their functions hinge on partnerships with two proteins that are redundantly required for stem cell maintenance. Here we focus on understanding how the essential partner protein, LST-1, modulates mRNA regulation by the PUF protein, FBF-2. LST-1 contains two nonidentical sites of interaction with FBF-2, LST-1 A and B. Our crystal structures of complexes of FBF-2, LST-1 A, and RNA visualize how FBF-2 associates with LST-1 A versus LST-1 B. One commonality is that FBF-2 contacts the conserved lysine and leucine side chains in the KxxL motifs in LST-1 A and B. A key difference is that FBF-2 forms unique contacts with regions N- and C-terminal to the KxxL motif. Consequently, LST-1 A does not modulate the RNA-binding affinity of FBF-2, whereas LST-1 B decreases RNA-binding affinity of FBF-2. The N-terminal region of LST-1 B, which binds near the 5' end of RNA elements, is essential to modulate FBF-2 RNA-binding affinity, while the C-terminal residues of LST-1 B contribute strong binding affinity to FBF-2. We conclude that LST-1 has the potential to impact which mRNAs are regulated depending on the precise nature of engagement through its functionally distinct FBF binding sites.