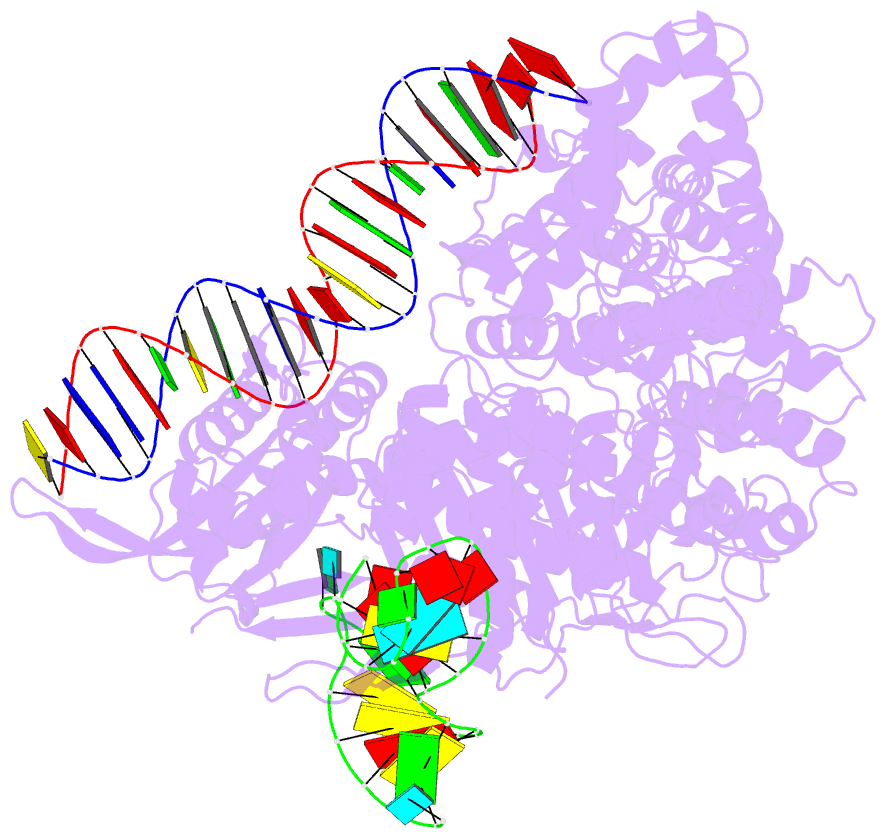
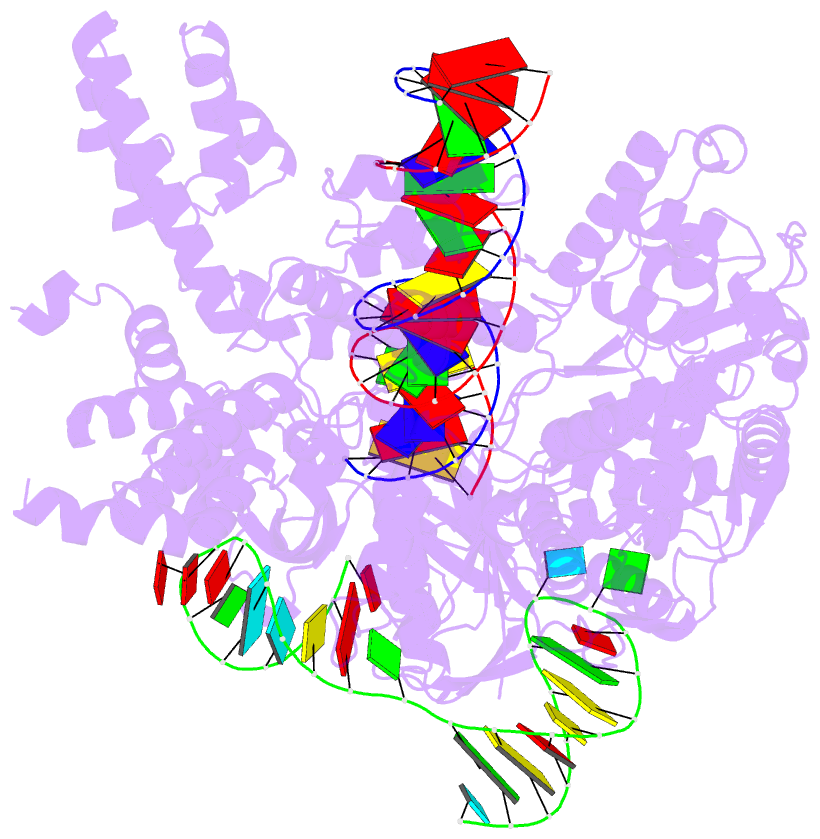
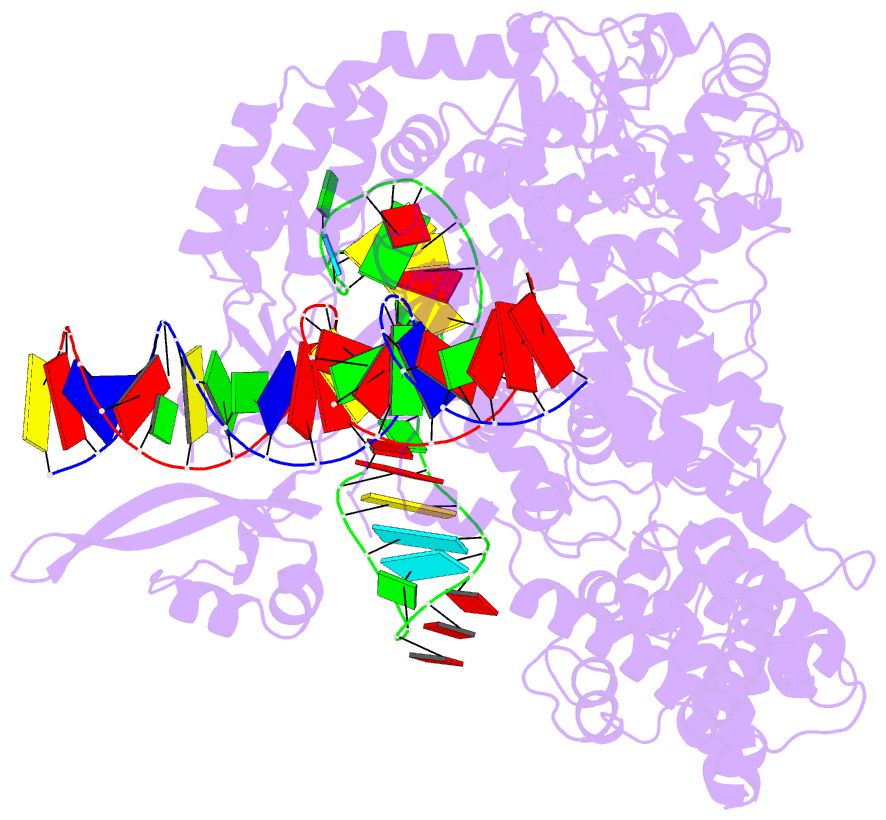
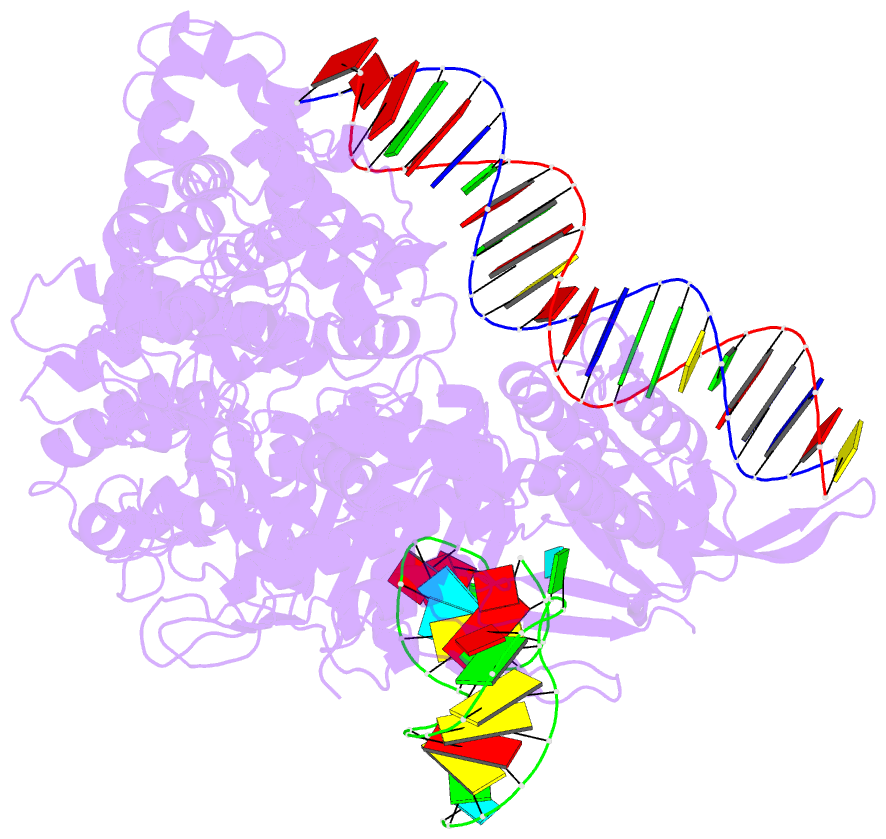
Summary information and primary citation
- PDB-id
- 7s3h; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-RNA-DNA
- Method
- cryo-EM (2.5 Å)
- Summary
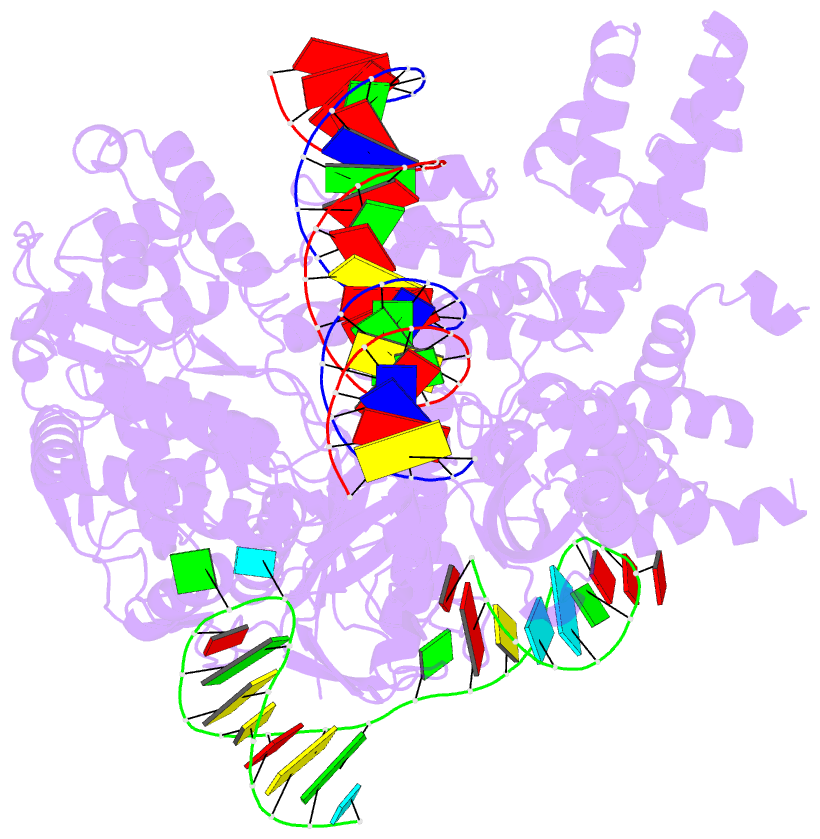
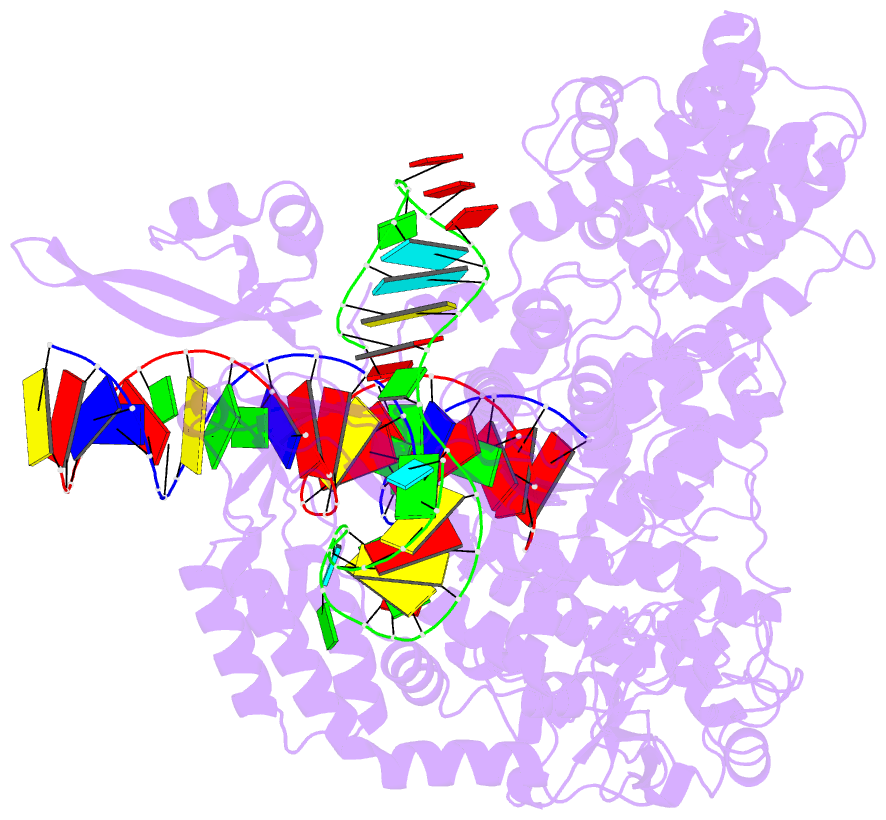
- Cas9:sgrna:DNA (s. pyogenes) with 0 RNA:DNA base pairs, open-protein-linear-DNA conformation
- Reference
- Cofsky JC, Soczek KM, Knott GJ, Nogales E, Doudna JA (2022): "CRISPR-Cas9 bends and twists DNA to read its sequence." Nat.Struct.Mol.Biol., 29, 395-402. doi: 10.1038/s41594-022-00756-0.
- Abstract
- In bacterial defense and genome editing applications, the CRISPR-associated protein Cas9 searches millions of DNA base pairs to locate a 20-nucleotide, guide RNA-complementary target sequence that abuts a protospacer-adjacent motif (PAM). Target capture requires Cas9 to unwind DNA at candidate sequences using an unknown ATP-independent mechanism. Here we show that Cas9 sharply bends and undertwists DNA on PAM binding, thereby flipping DNA nucleotides out of the duplex and toward the guide RNA for sequence interrogation. Cryogenic-electron microscopy (cryo-EM) structures of Cas9-RNA-DNA complexes trapped at different states of the interrogation pathway, together with solution conformational probing, reveal that global protein rearrangement accompanies formation of an unstacked DNA hinge. Bend-induced base flipping explains how Cas9 'reads' snippets of DNA to locate target sites within a vast excess of nontarget DNA, a process crucial to both bacterial antiviral immunity and genome editing. This mechanism establishes a physical solution to the problem of complementarity-guided DNA search and shows how interrogation speed and local DNA geometry may influence genome editing efficiency.