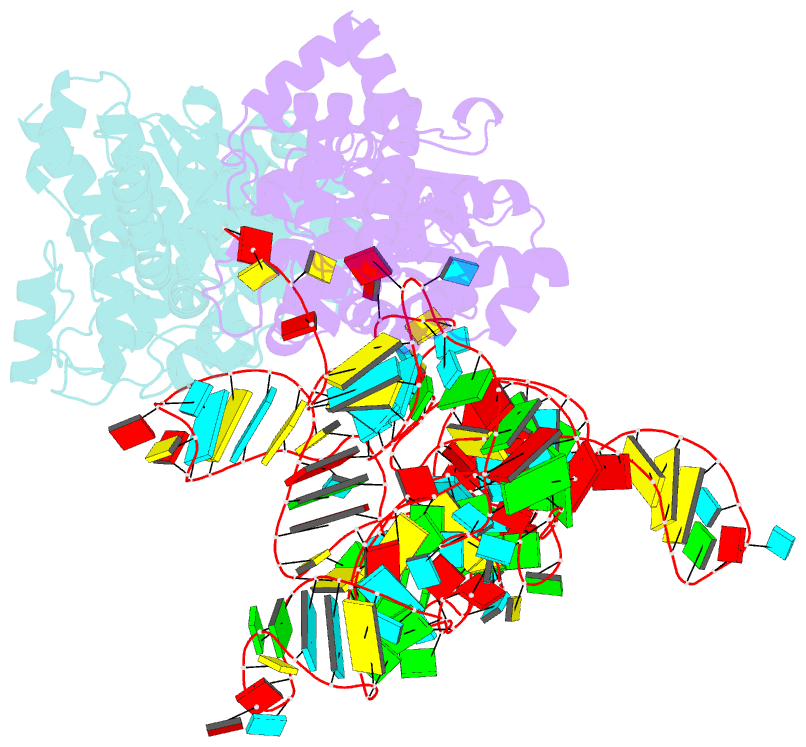
Summary information and primary citation
- PDB-id
- 7scq; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- cryo-EM (6.0 Å)
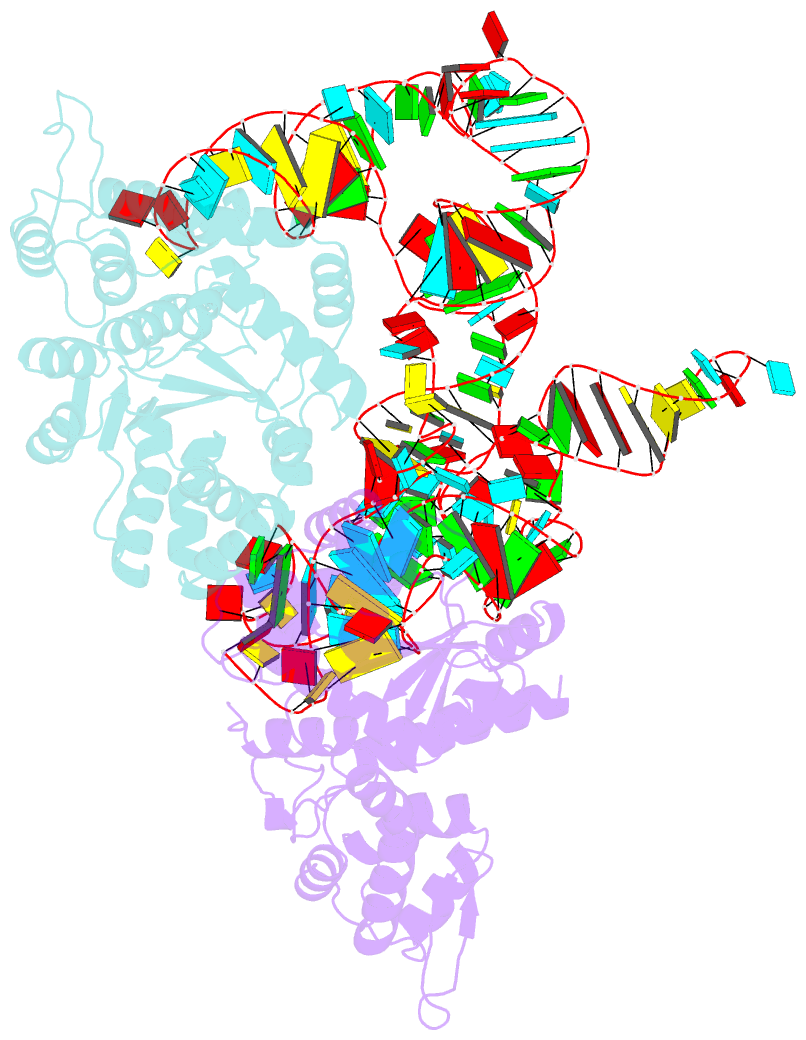
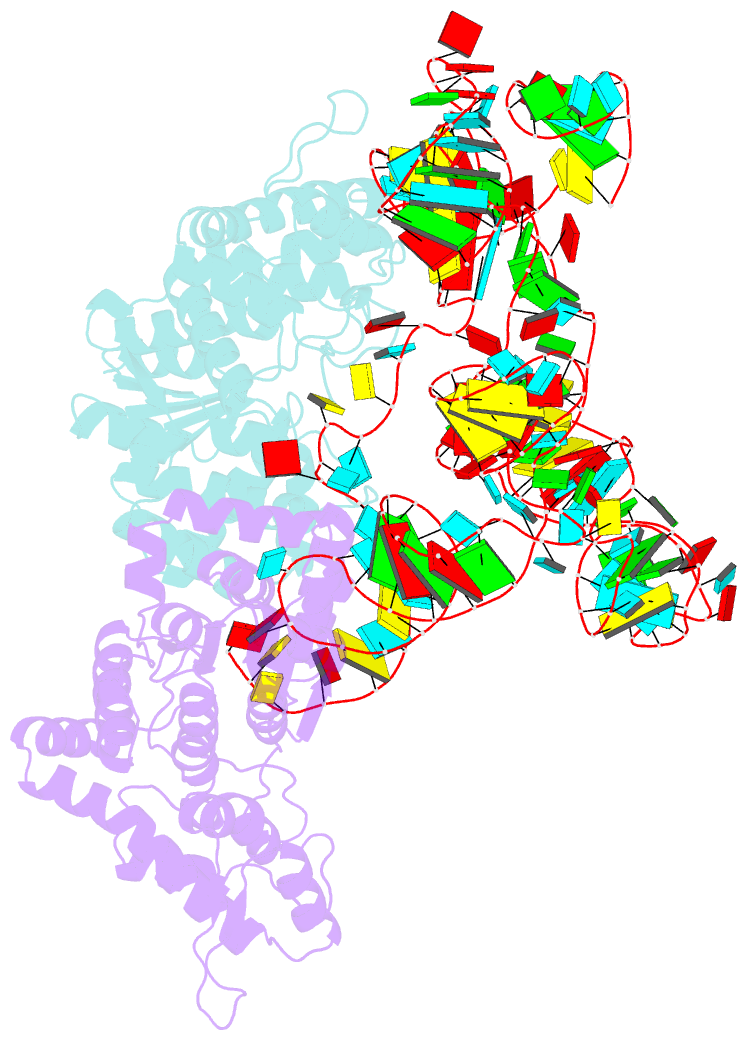
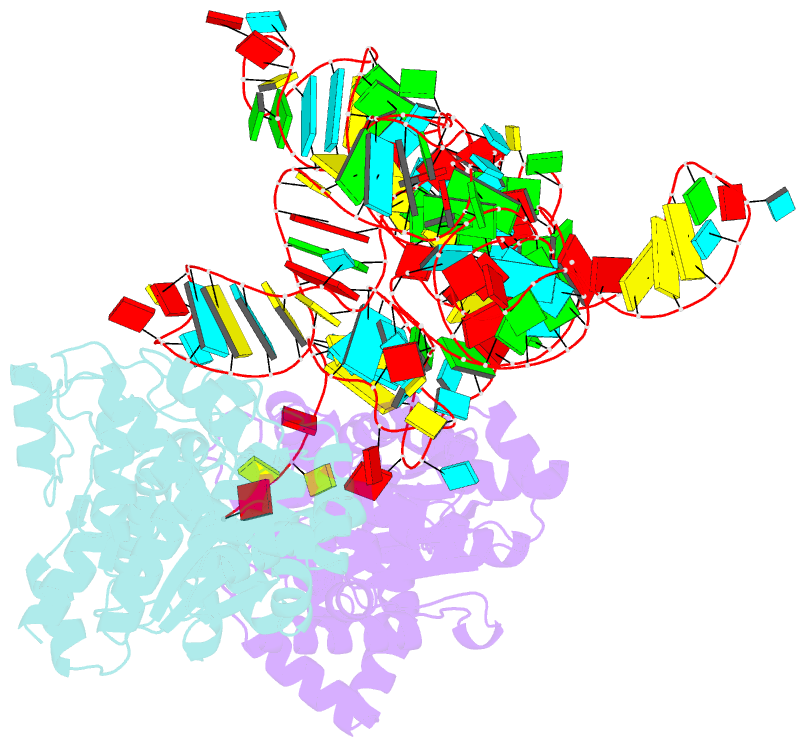
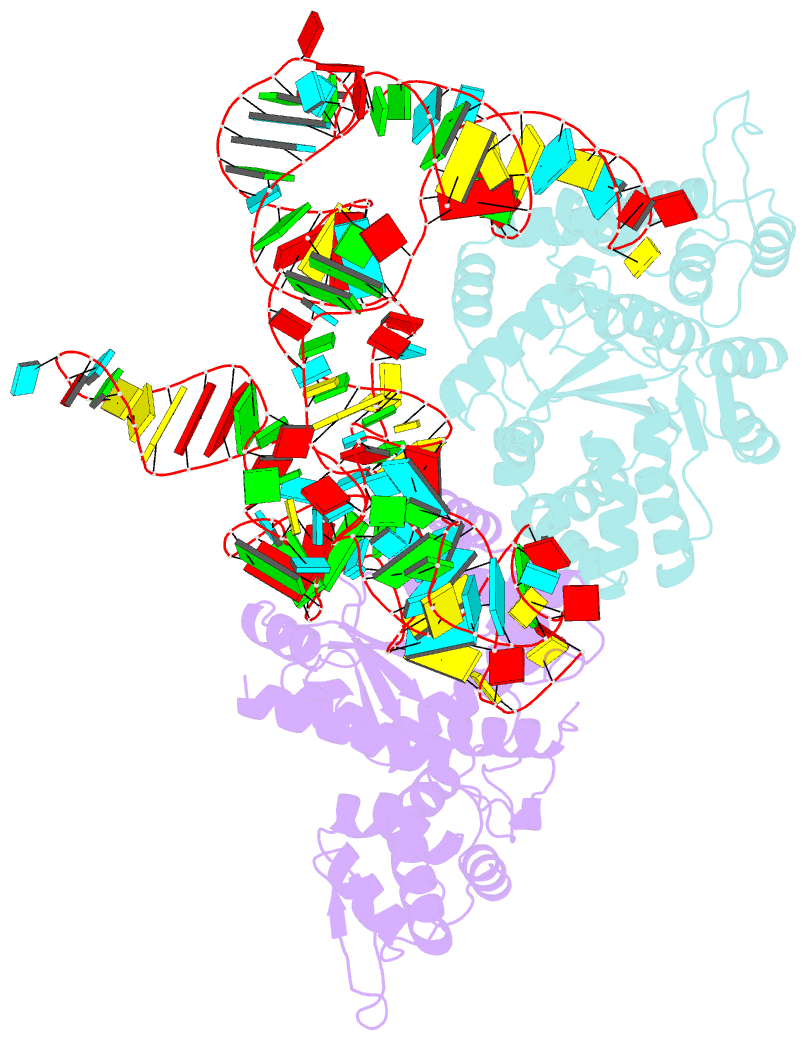
- Summary
- Trna-like structure from brome mosaic virus bound to tyrosyl-trna synthetase from phaseolus vulgaris. conformation: bound state 2.
- Reference
- Bonilla SL, Sherlock ME, MacFadden A, Kieft JS (2021): "A viral RNA hijacks host machinery using dynamic conformational changes of a tRNA-like structure." Science, 374, 955-960. doi: 10.1126/science.abe8526.
- Abstract
- Viruses require multifunctional structured RNAs to hijack their host’s biochemistry, but their mechanisms can be obscured by the difficulty of solving conformationally dynamic RNA structures. Using cryo–electron microscopy (cryo-EM), we visualized the structure of the mysterious viral transfer RNA (tRNA)–like structure (TLS) from the brome mosaic virus, which affects replication, translation, and genome encapsidation. Structures in isolation and those bound to tyrosyl-tRNA synthetase (TyrRS) show that this ~55-kilodalton purported tRNA mimic undergoes large conformational rearrangements to bind TyrRS in a form that differs substantially from that of tRNA. Our study reveals how viral RNAs can use a combination of static and dynamic RNA structures to bind host machinery through highly noncanonical interactions, and we highlight the utility of cryo-EM for visualizing small, conformationally dynamic structured RNAs.