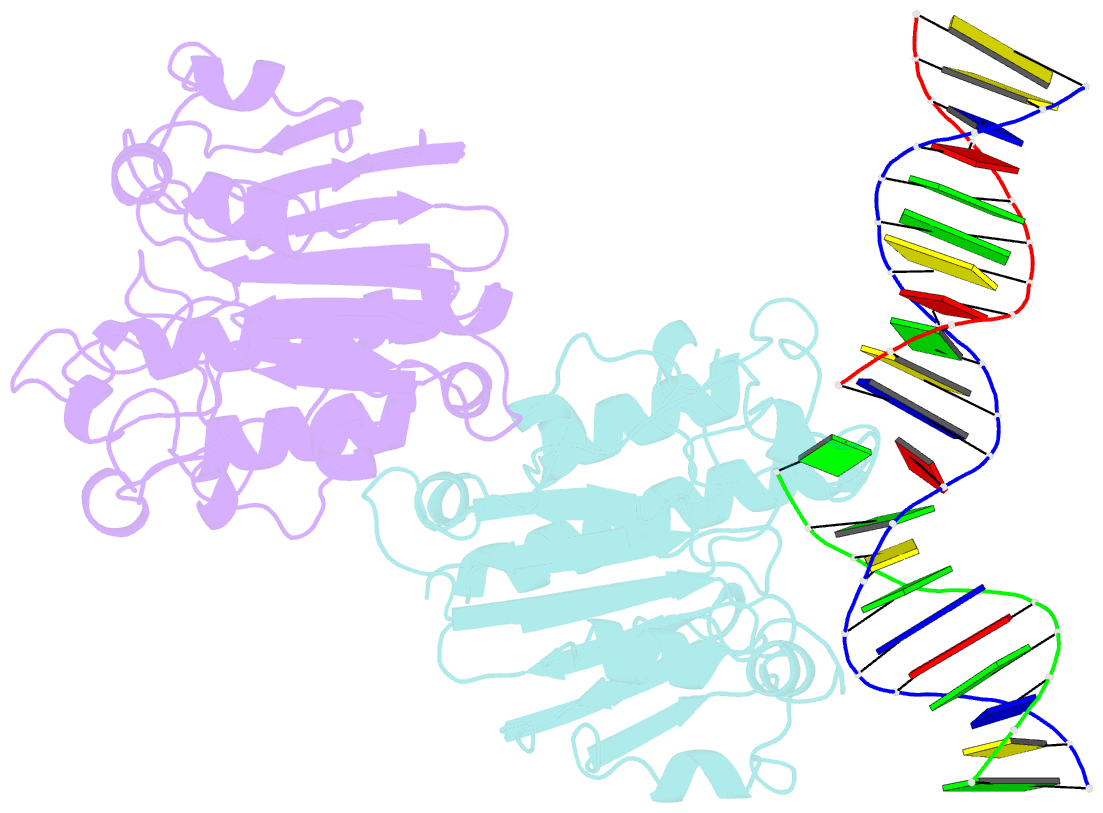
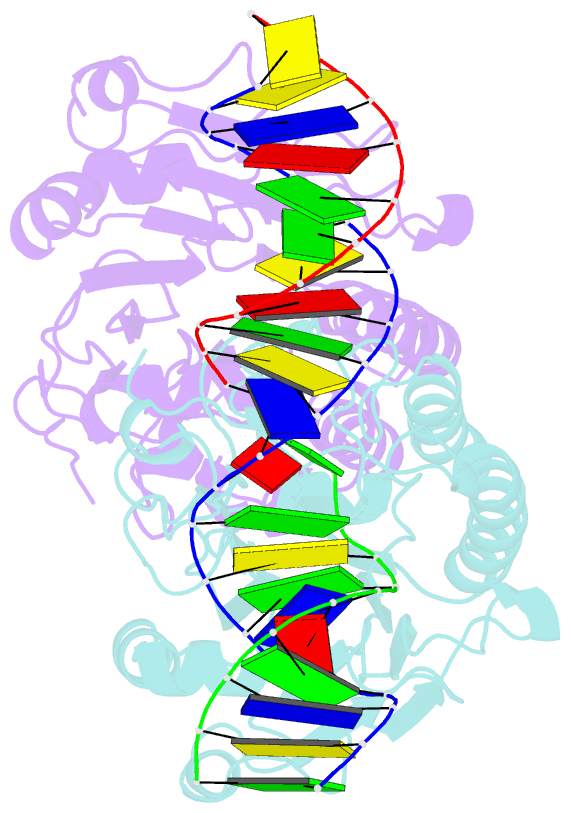
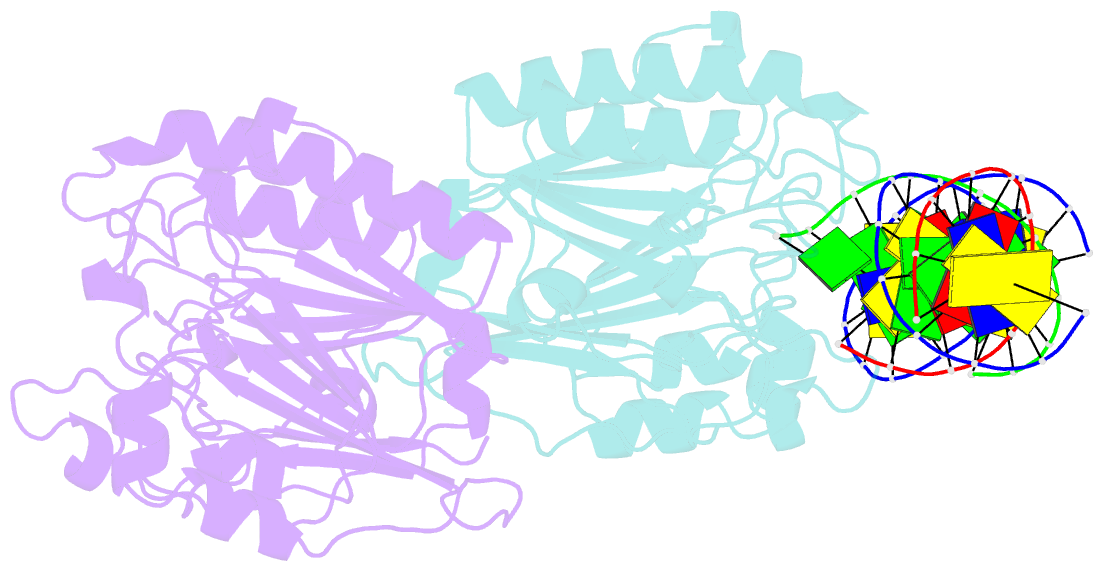
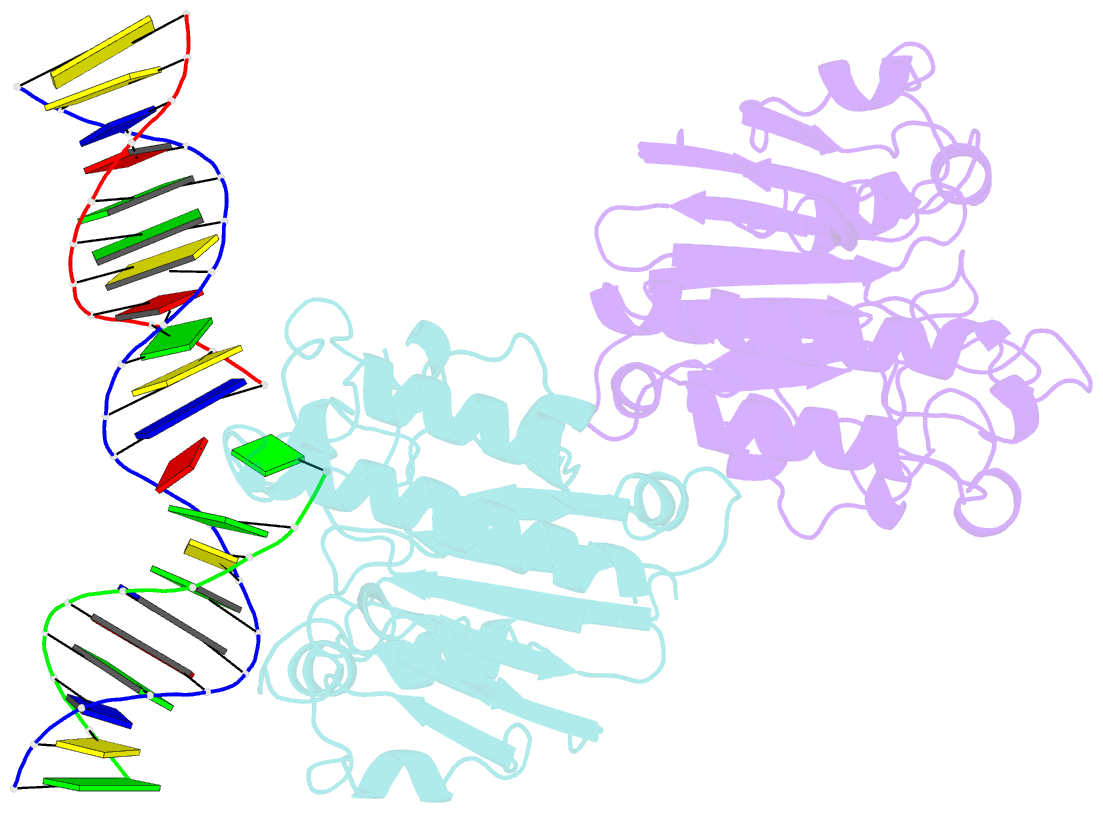
Summary information and primary citation
- PDB-id
- 7suv; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (1.99 Å)
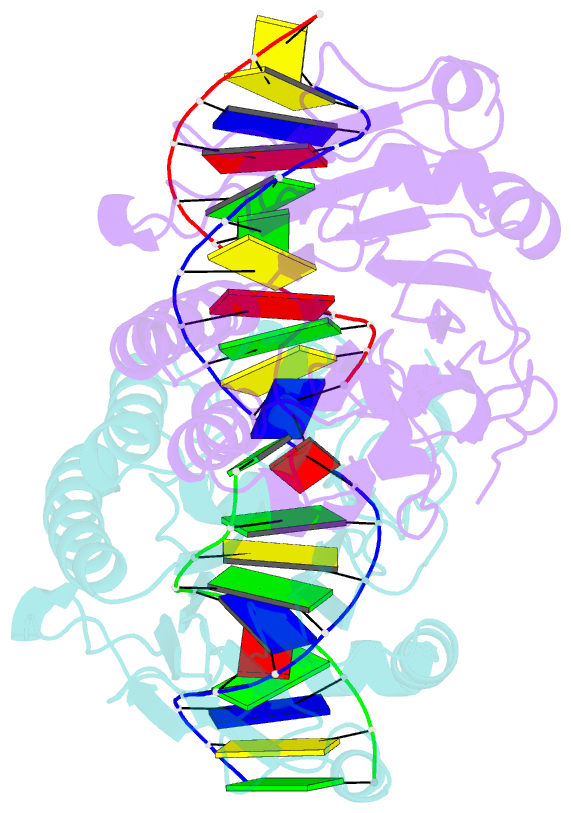
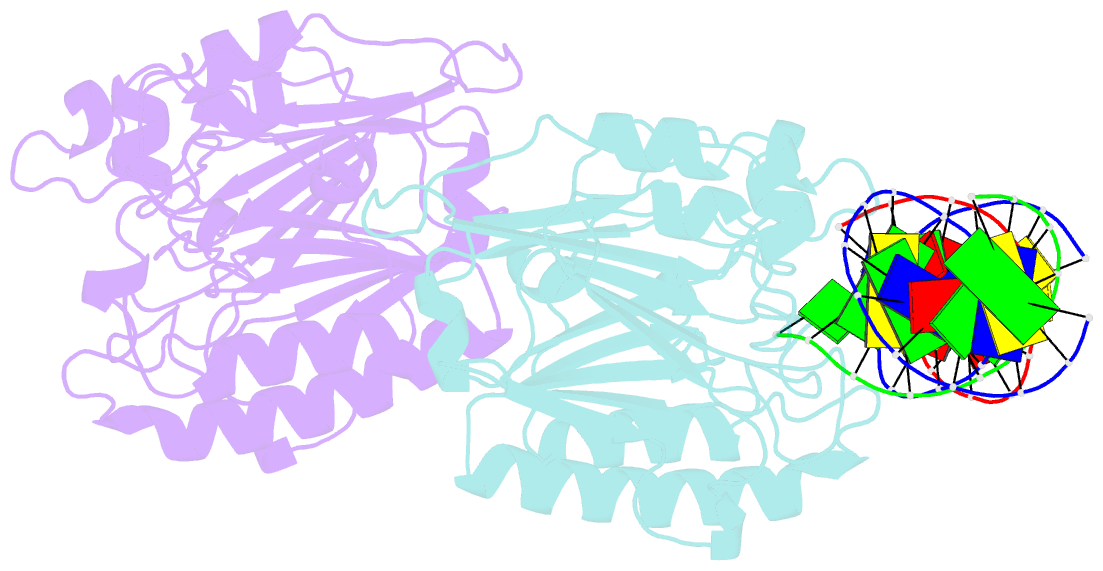
- Summary
- Ape1 exonuclease substrate complex with 8oxog opposite a
- Reference
- Whitaker AM, Stark WJ, Freudenthal BD (2022): "Processing oxidatively damaged bases at DNA strand breaks by APE1." Nucleic Acids Res., 50, 9521-9533. doi: 10.1093/nar/gkac695.
- Abstract
- Reactive oxygen species attack the structure of DNA, thus altering its base-pairing properties. Consequently, oxidative stress-associated DNA lesions are a major source of the mutation load that gives rise to cancer and other diseases. Base excision repair (BER) is the pathway primarily tasked with repairing DNA base damage, with apurinic/apyrimidinic endonuclease (APE1) having both AP-endonuclease and 3' to 5' exonuclease (exo) DNA cleavage functions. The lesion 8-oxo-7,8-dihydroguanine (8-oxoG) can enter the genome as either a product of direct damage to the DNA, or through polymerase insertion at the 3'-end of a DNA strand during replication or repair. Importantly, 3'-8-oxoG impairs the ligation step of BER and therefore must be removed by the exo activity of a surrogate enzyme to prevent double stranded breaks and cell death. In the present study, we use X-ray crystallography to characterize the exo activity of APE1 on 3'-8-oxoG substrates. These structures support a unified APE1 exo mechanism that differs from its more canonical AP-endonuclease activity. In addition, through complementation of the structural data with enzyme kinetics and binding studies employing both wild-type and rationally designed APE1 mutants, we were able to identify and characterize unique protein: DNA contacts that specifically mediate 8-oxoG removal by APE1.