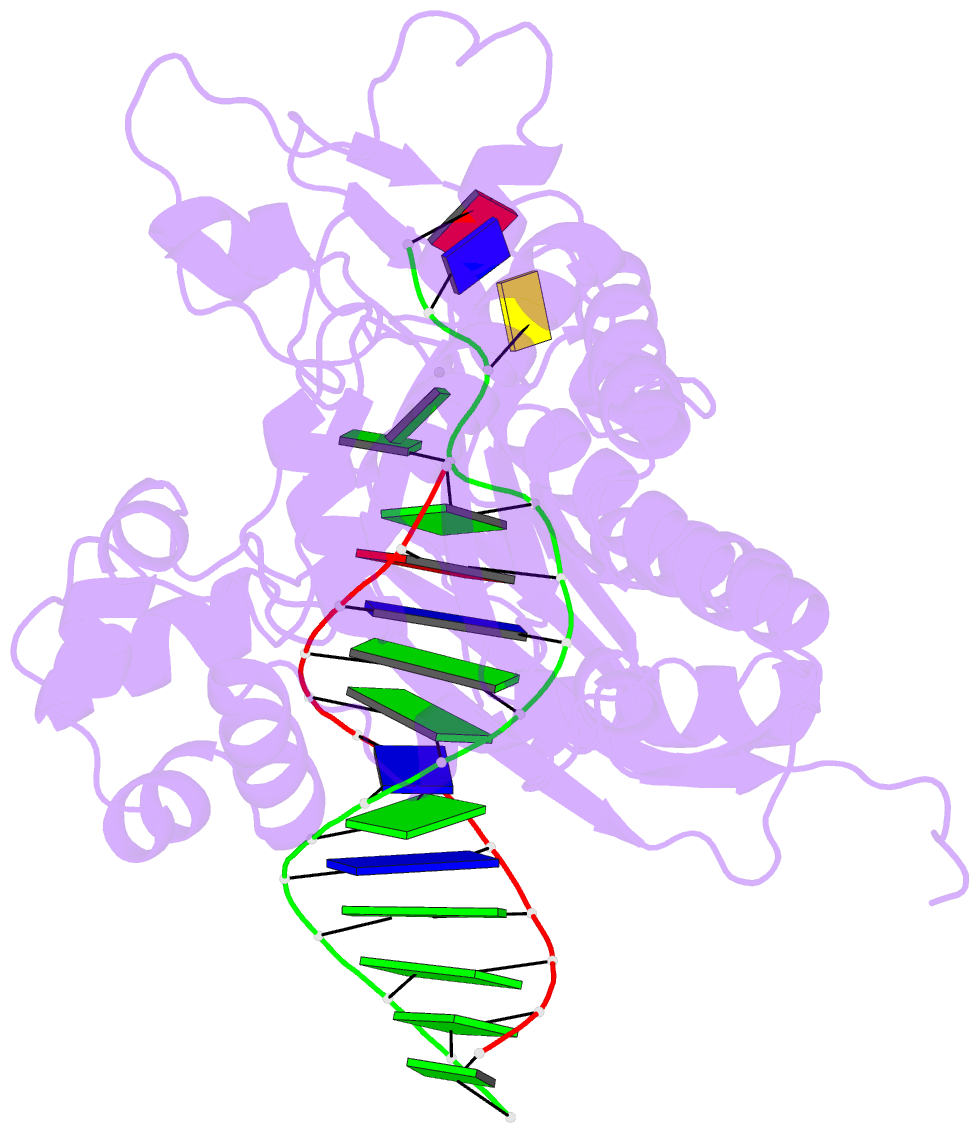
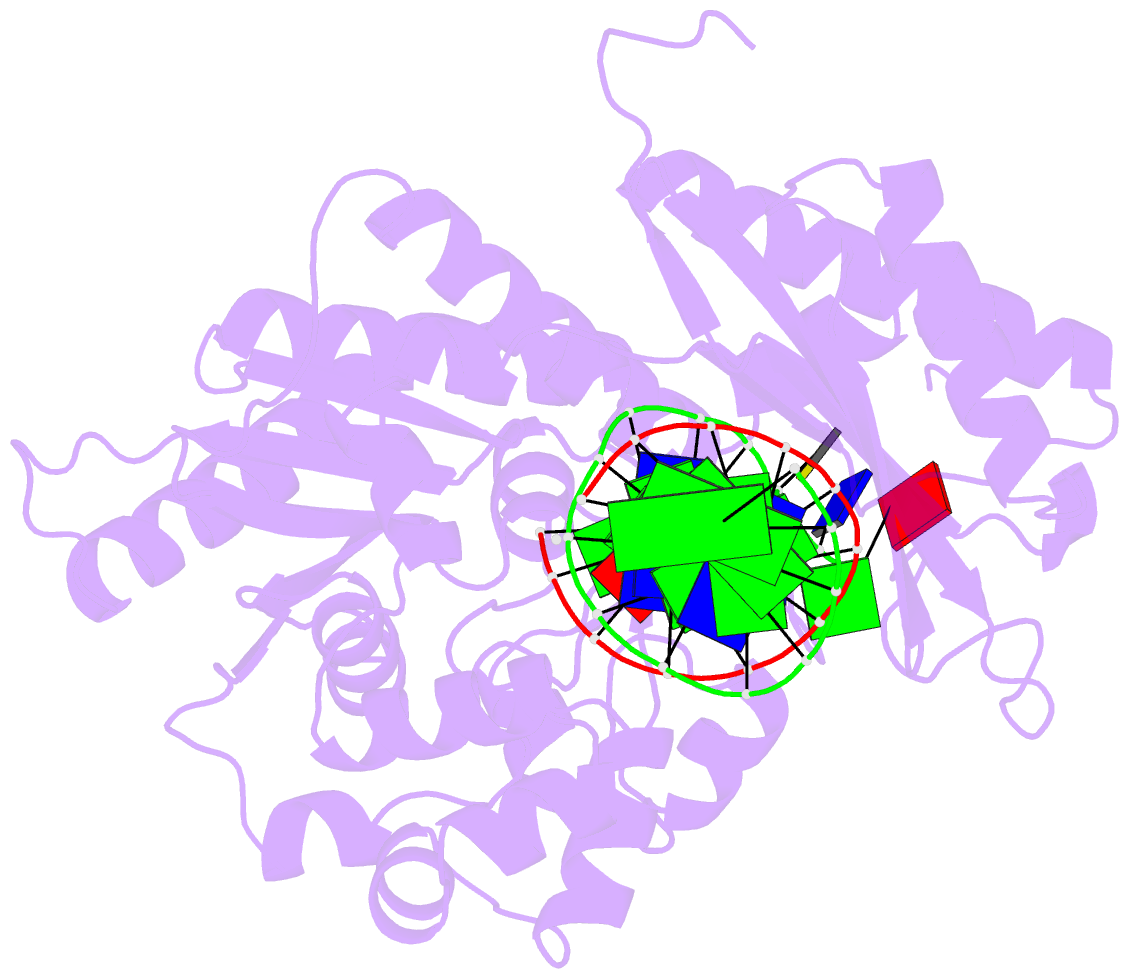
Summary information and primary citation
- PDB-id
- 7t19; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.01 Å)
- Summary
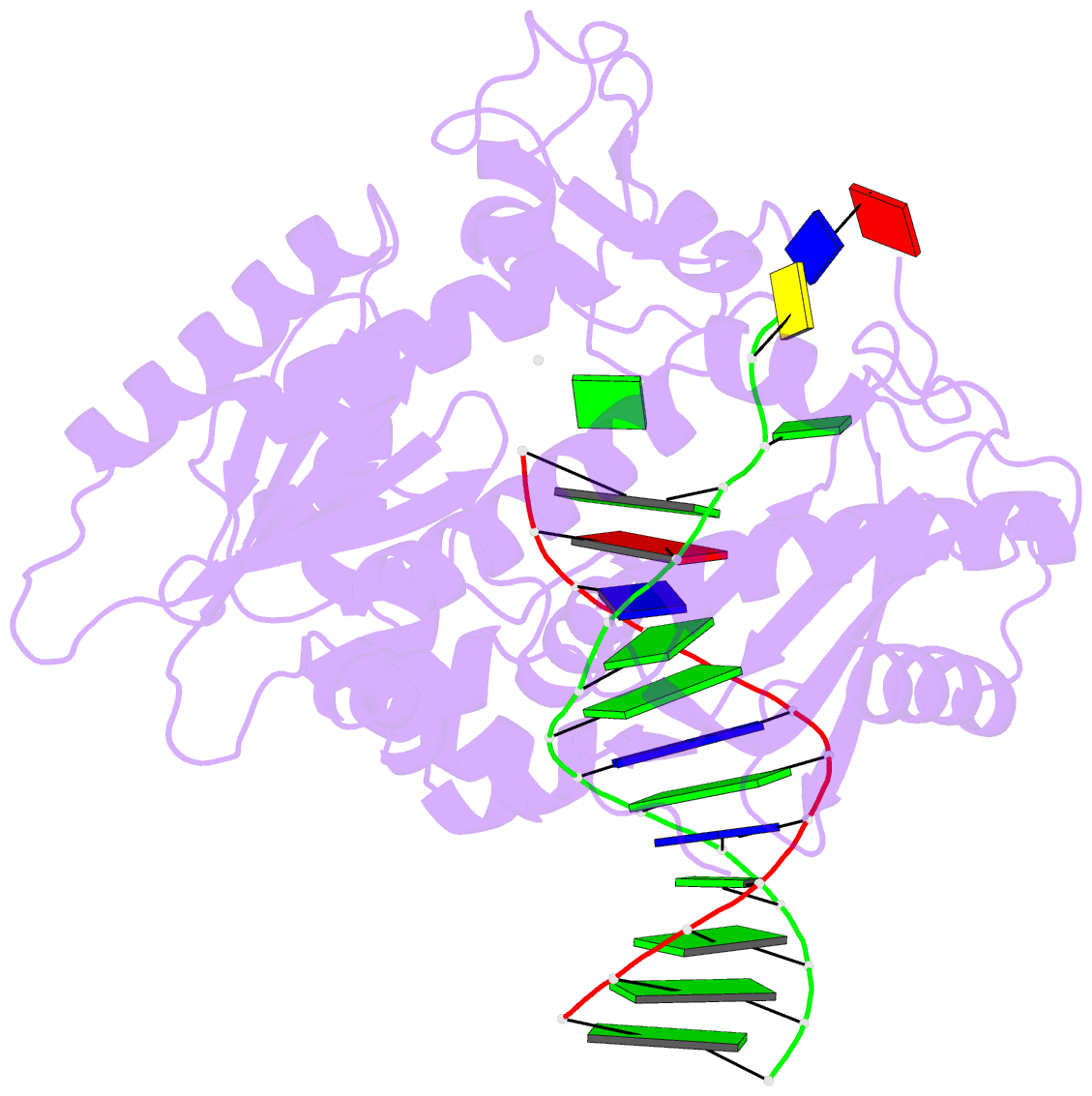
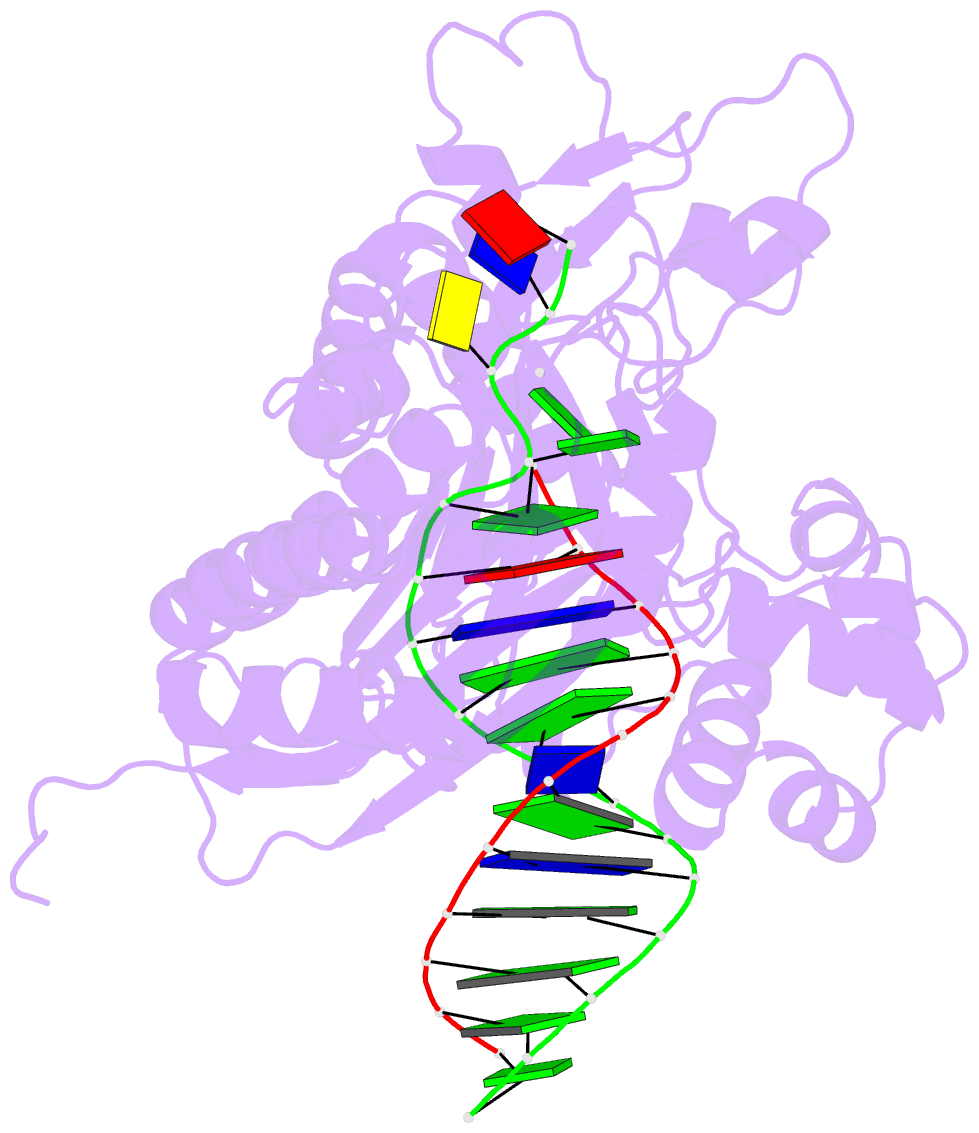
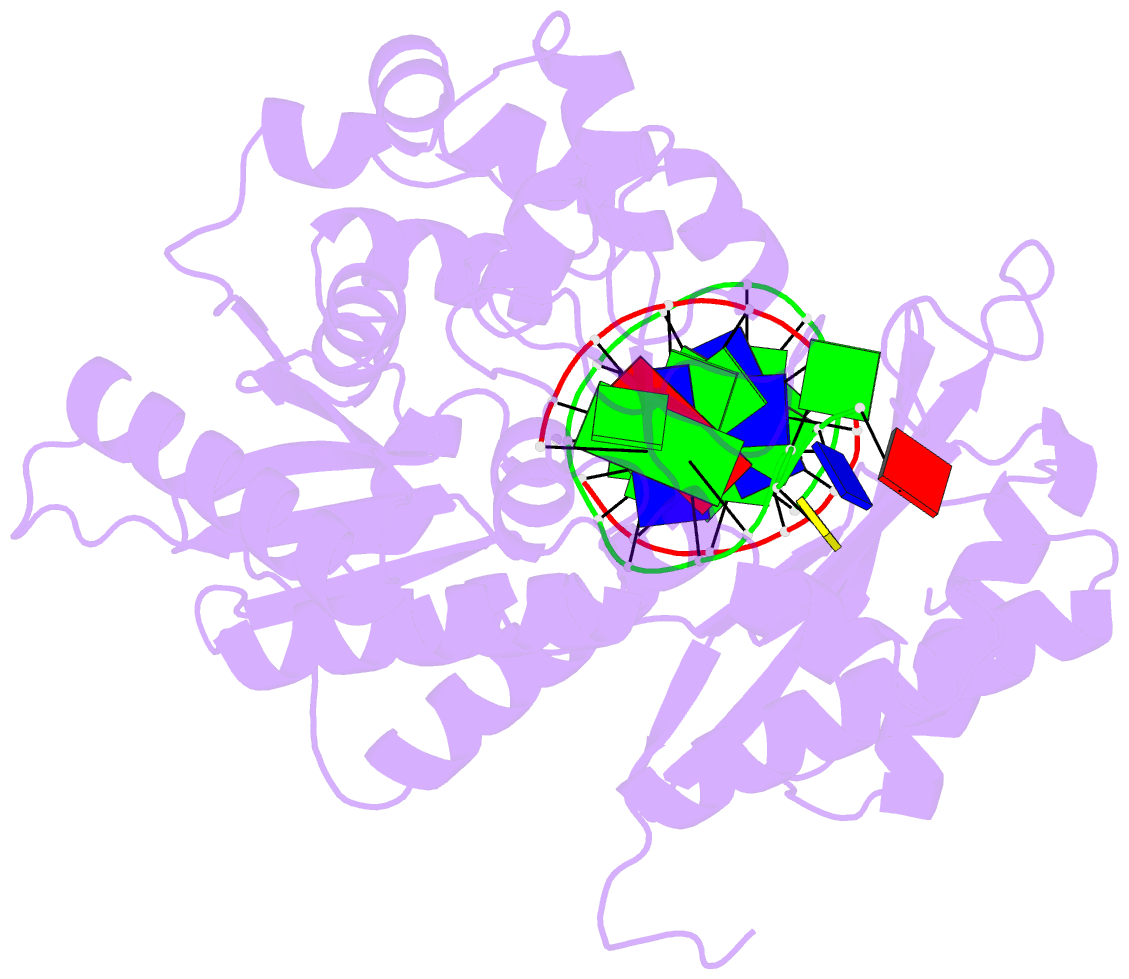
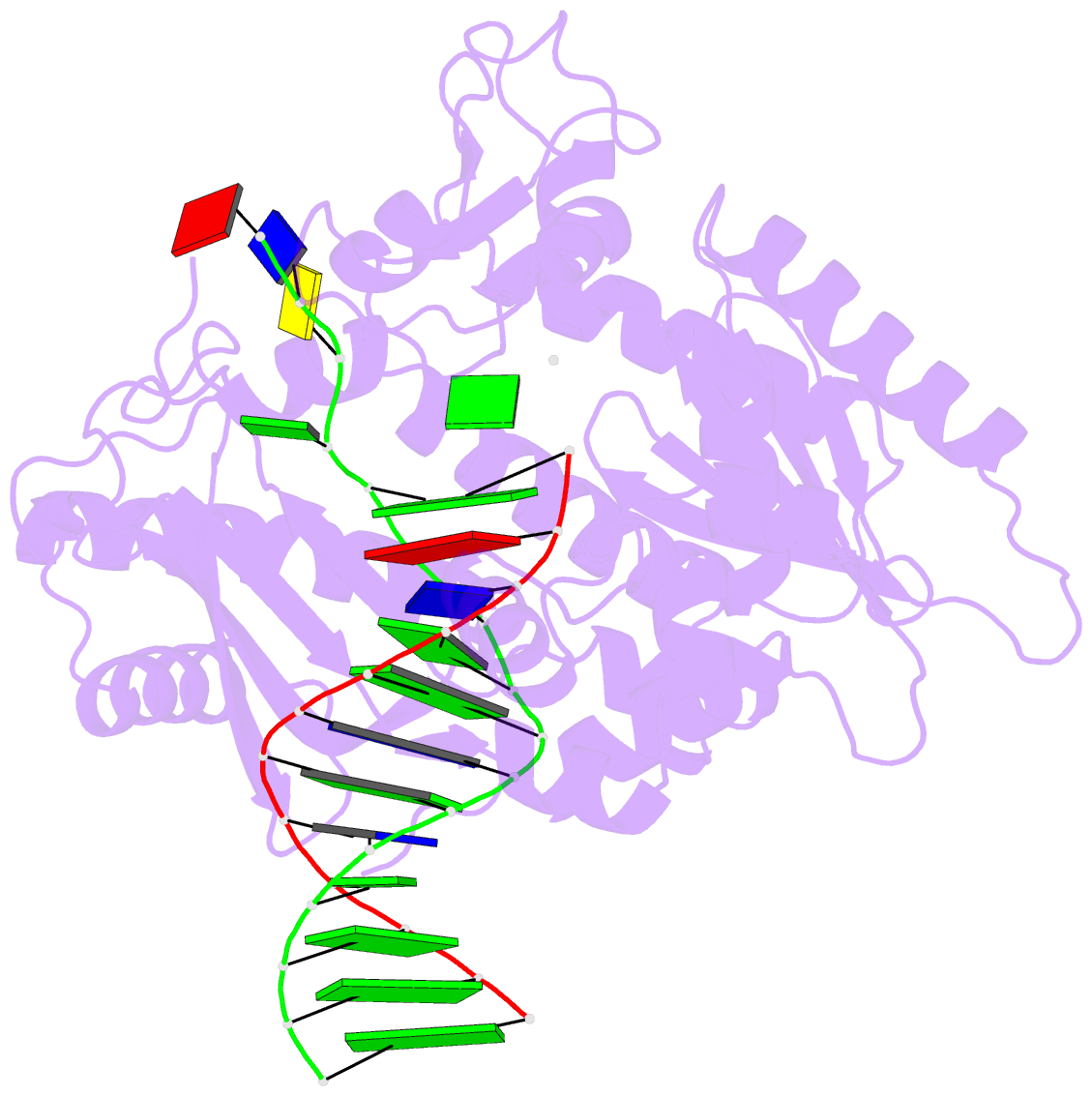
- Rev1 ternary complex with dgtp and ca2+
- Reference
- Weaver TM, Click TH, Khoang TH, Todd Washington M, Agarwal PK, Freudenthal BD (2022): "Mechanism of nucleotide discrimination by the translesion synthesis polymerase Rev1." Nat Commun, 13, 2876. doi: 10.1038/s41467-022-30577-0.
- Abstract
- Rev1 is a translesion DNA synthesis (TLS) polymerase involved in the bypass of adducted-guanine bases and abasic sites during DNA replication. During damage bypass, Rev1 utilizes a protein-template mechanism of DNA synthesis, where the templating DNA base is evicted from the Rev1 active site and replaced by an arginine side chain that preferentially binds incoming dCTP. Here, we utilize X-ray crystallography and molecular dynamics simulations to obtain structural insight into the dCTP specificity of Rev1. We show the Rev1 R324 protein-template forms sub-optimal hydrogen bonds with incoming dTTP, dGTP, and dATP that prevents Rev1 from adopting a catalytically competent conformation. Additionally, we show the Rev1 R324 protein-template forms optimal hydrogen bonds with incoming rCTP. However, the incoming rCTP adopts an altered sugar pucker, which prevents the formation of a catalytically competent Rev1 active site. This work provides novel insight into the mechanisms for nucleotide discrimination by the TLS polymerase Rev1.