Summary information and primary citation
- PDB-id
- 7tan; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- structural protein-DNA-transferase
- Method
- cryo-EM (3.0 Å)
- Summary
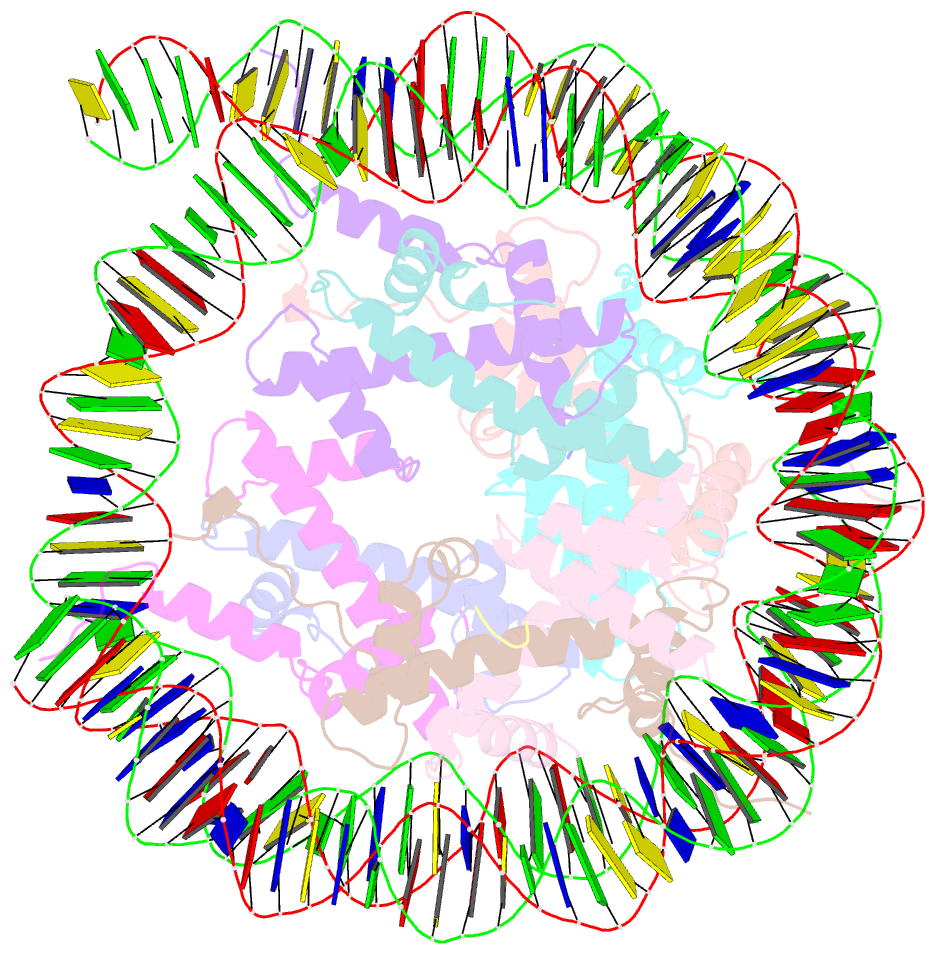
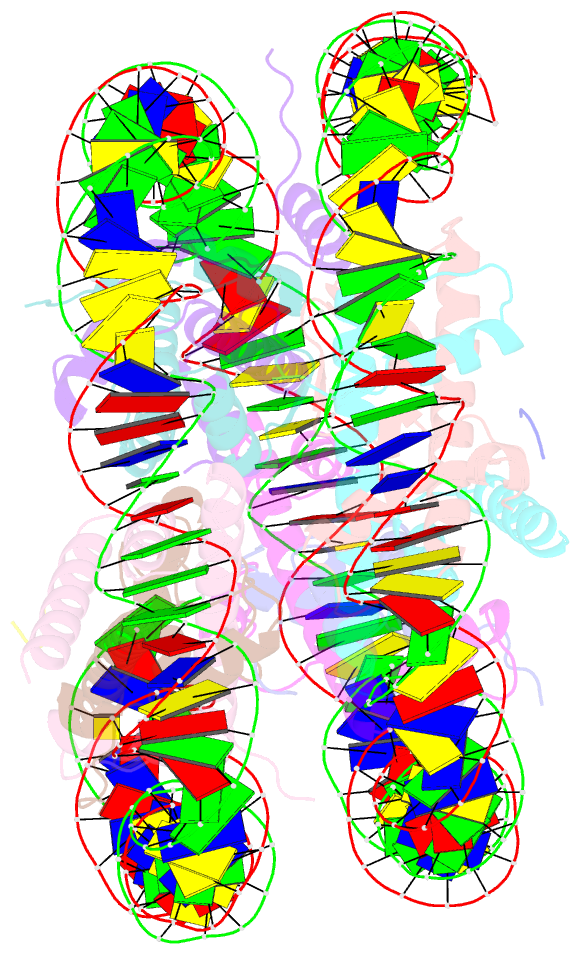
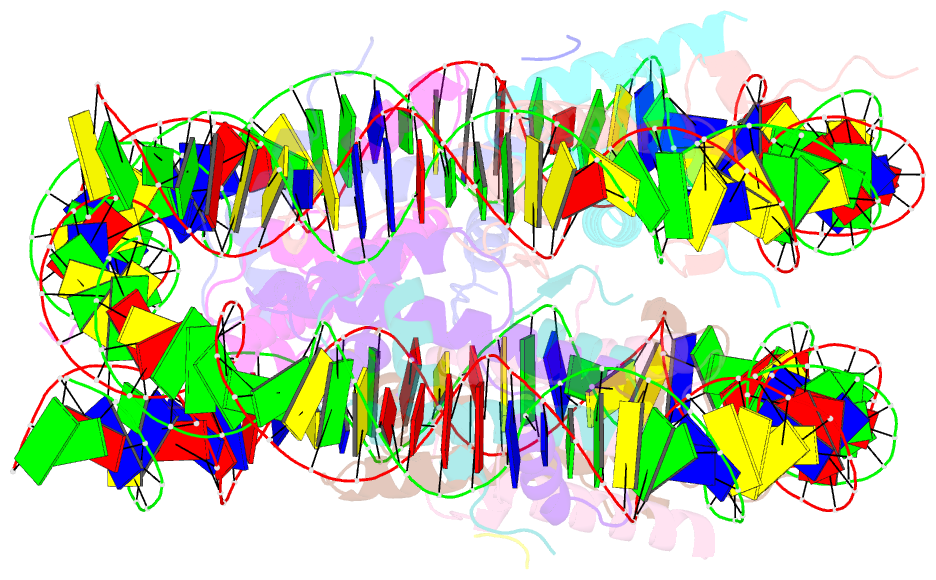
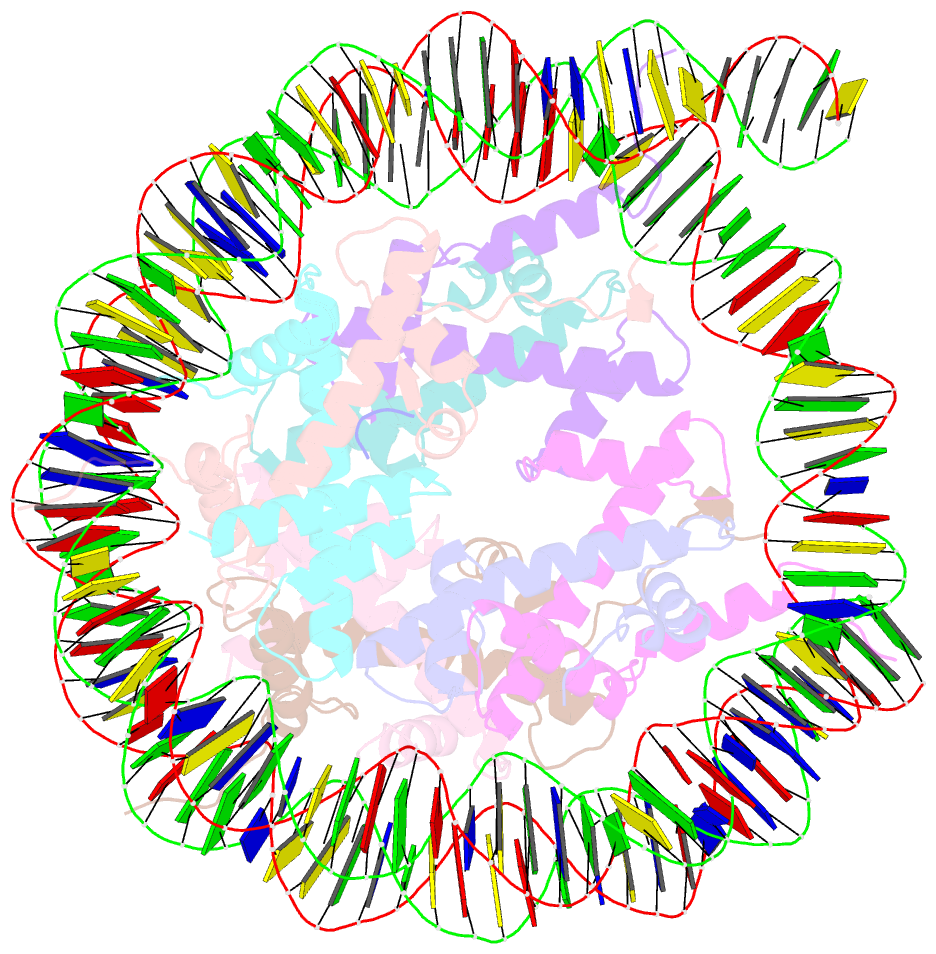
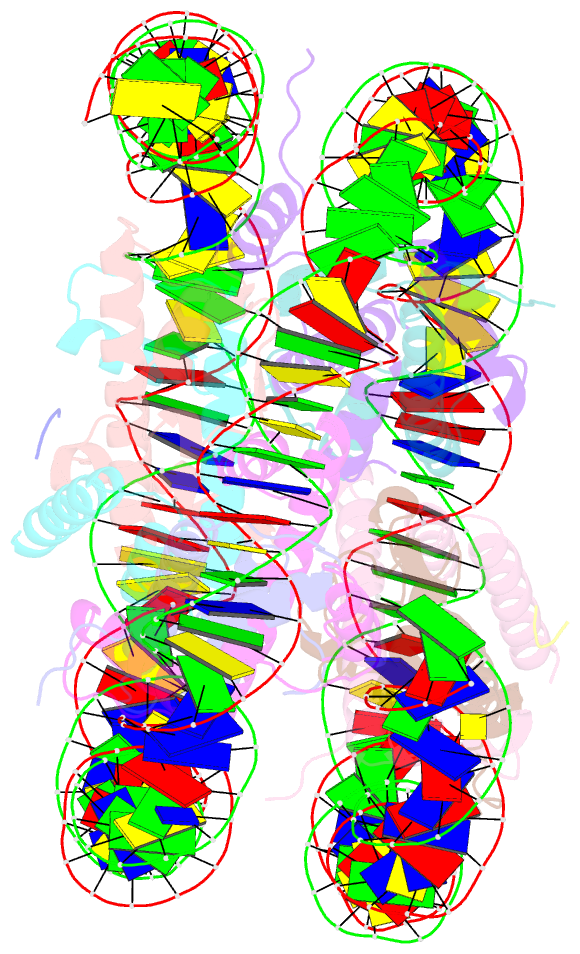
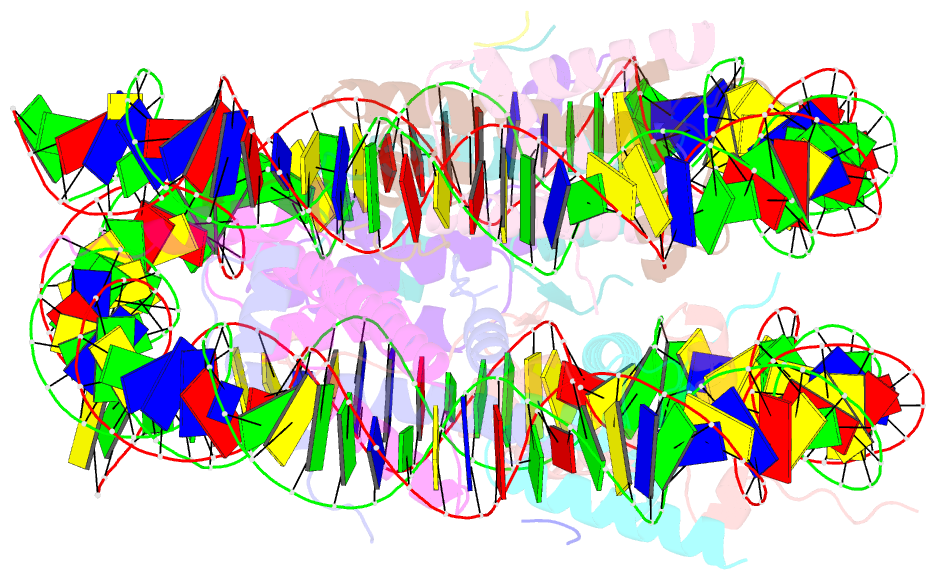
- Structure of vrk1 c-terminal tail bound to nucleosome core particle
- Reference
- Budziszewski GR, Zhao Y, Spangler CJ, Kedziora KM, Williams MR, Azzam DN, Skrajna A, Koyama Y, Cesmat AP, Simmons HC, Arteaga EC, Strauss JD, Kireev D, McGinty RK (2022): "Multivalent DNA and nucleosome acidic patch interactions specify VRK1 mitotic localization and activity." Nucleic Acids Res., 50, 4355-4371. doi: 10.1093/nar/gkac198.
- Abstract
- A key role of chromatin kinases is to phosphorylate histone tails during mitosis to spatiotemporally regulate cell division. Vaccinia-related kinase 1 (VRK1) is a serine-threonine kinase that phosphorylates histone H3 threonine 3 (H3T3) along with other chromatin-based targets. While structural studies have defined how several classes of histone-modifying enzymes bind to and function on nucleosomes, the mechanism of chromatin engagement by kinases is largely unclear. Here, we paired cryo-electron microscopy with biochemical and cellular assays to demonstrate that VRK1 interacts with both linker DNA and the nucleosome acidic patch to phosphorylate H3T3. Acidic patch binding by VRK1 is mediated by an arginine-rich flexible C-terminal tail. Homozygous missense and nonsense mutations of this acidic patch recognition motif in VRK1 are causative in rare adult-onset distal spinal muscular atrophy. We show that these VRK1 mutations interfere with nucleosome acidic patch binding, leading to mislocalization of VRK1 during mitosis, thus providing a potential new molecular mechanism for pathogenesis.