Summary information and primary citation
- PDB-id
- 7tdx; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (3.1 Å)
- Summary
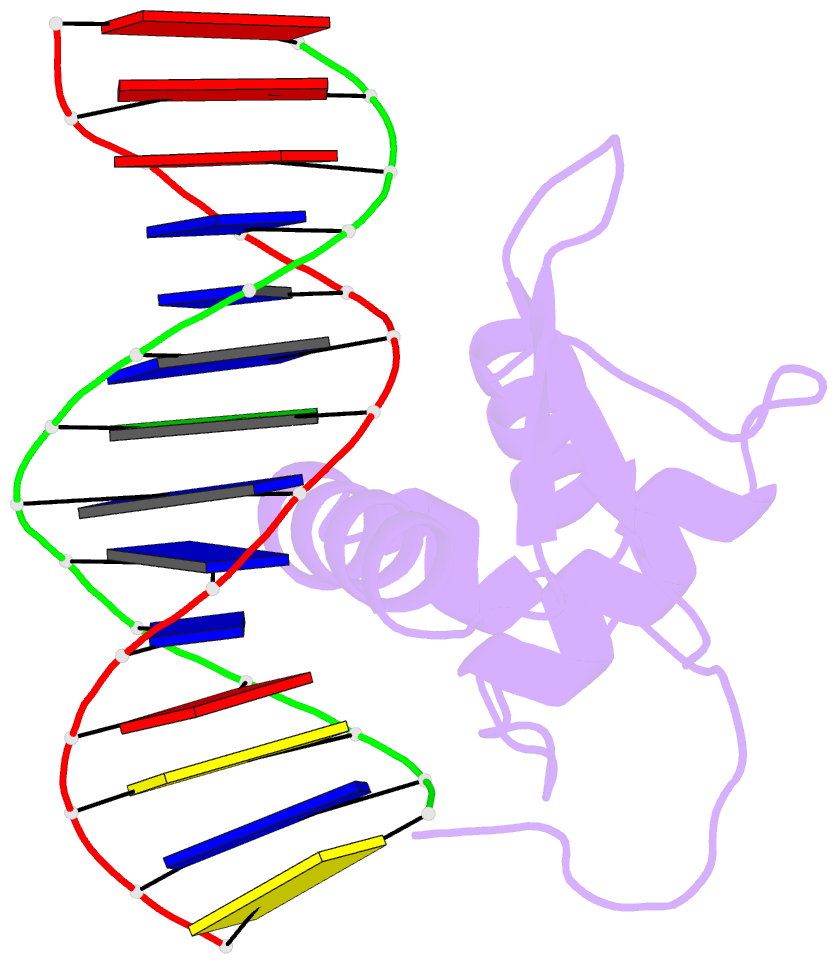
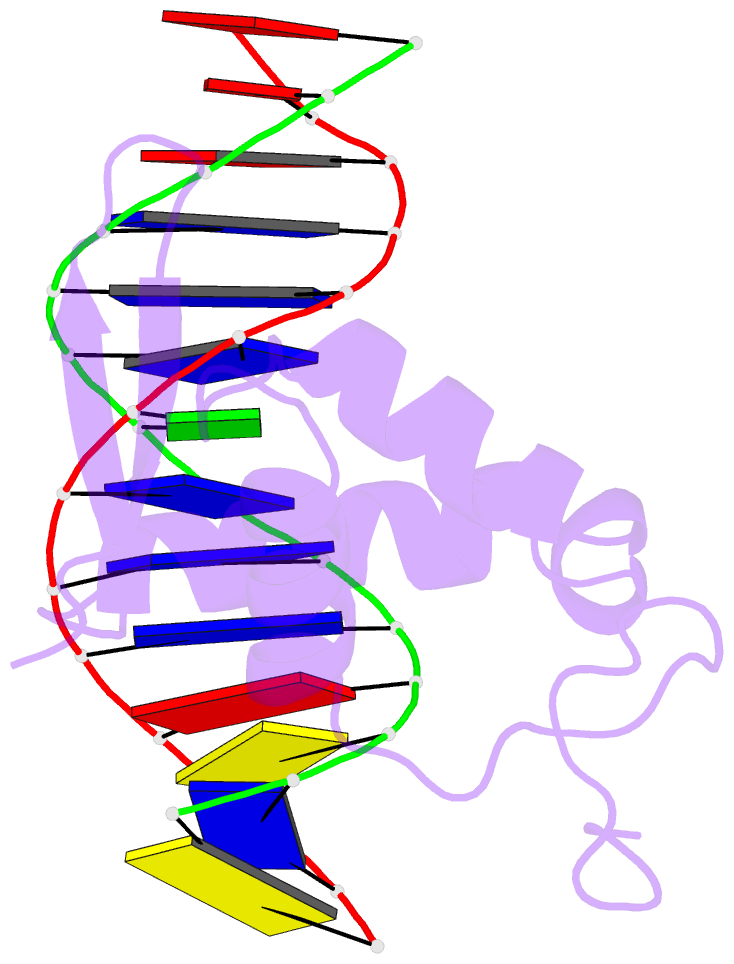
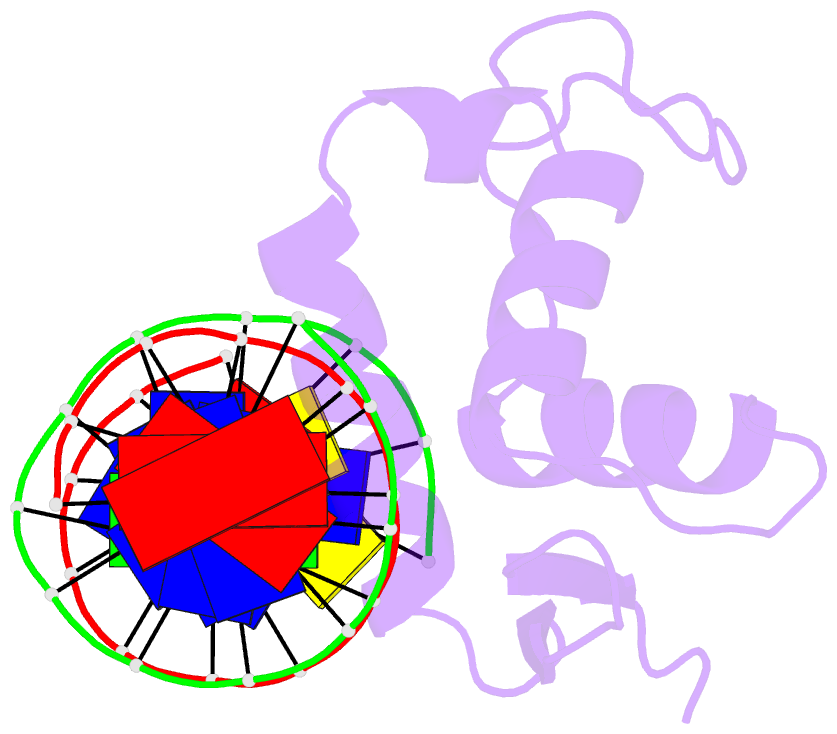
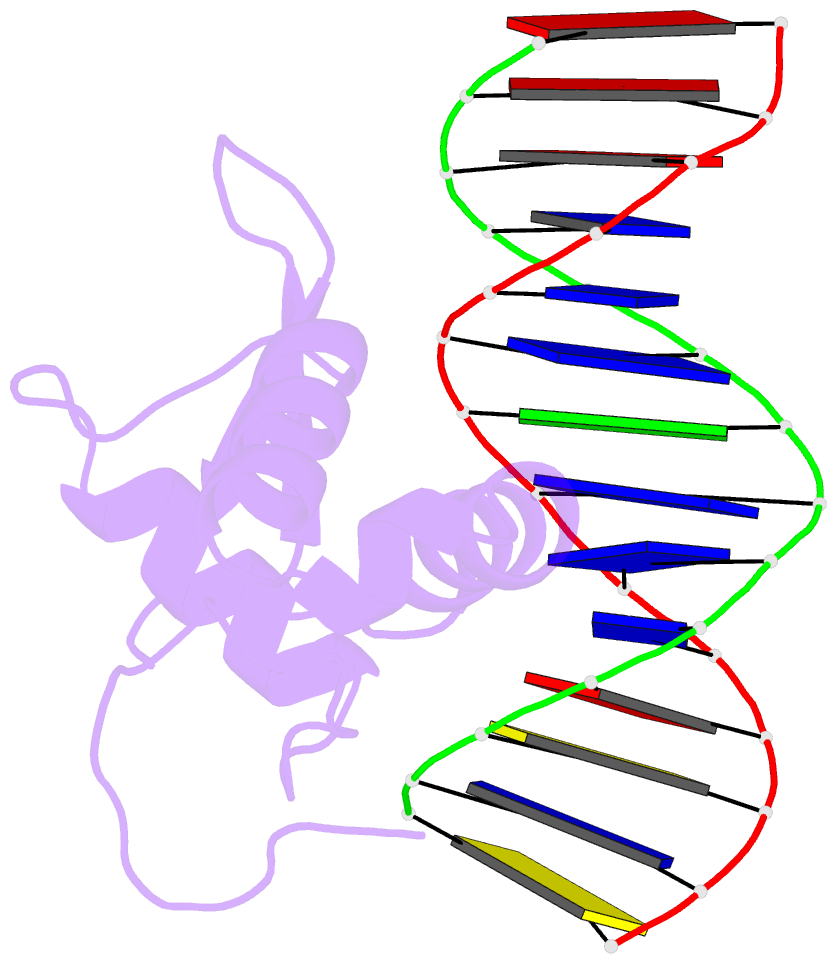
- Structure of foxp3-DNA complex
- Reference
- Leng F, Zhang W, Ramirez RN, Leon J, Zhong Y, Hou L, Yuki K, van der Veeken J, Rudensky AY, Benoist C, Hur S (2022): "The transcription factor FoxP3 can fold into two dimerization states with divergent implications for regulatory T cell function and immune homeostasis." Immunity, 55, 1354. doi: 10.1016/j.immuni.2022.07.002.
- Abstract
- FoxP3 is an essential transcription factor (TF) for immunologic homeostasis, but how it utilizes the common forkhead DNA-binding domain (DBD) to perform its unique function remains poorly understood. We here demonstrated that unlike other known forkhead TFs, FoxP3 formed a head-to-head dimer using a unique linker (Runx1-binding region [RBR]) preceding the forkhead domain. Head-to-head dimerization conferred distinct DNA-binding specificity and created a docking site for the cofactor Runx1. RBR was also important for proper folding of the forkhead domain, as truncation of RBR induced domain-swap dimerization of forkhead, which was previously considered the physiological form of FoxP3. Rather, swap-dimerization impaired FoxP3 function, as demonstrated with the disease-causing mutation R337Q, whereas a swap-suppressive mutation largely rescued R337Q-mediated functional impairment. Altogether, our findings suggest that FoxP3 can fold into two distinct dimerization states: head-to-head dimerization representing functional specialization of an ancient DBD and swap dimerization associated with impaired functions.