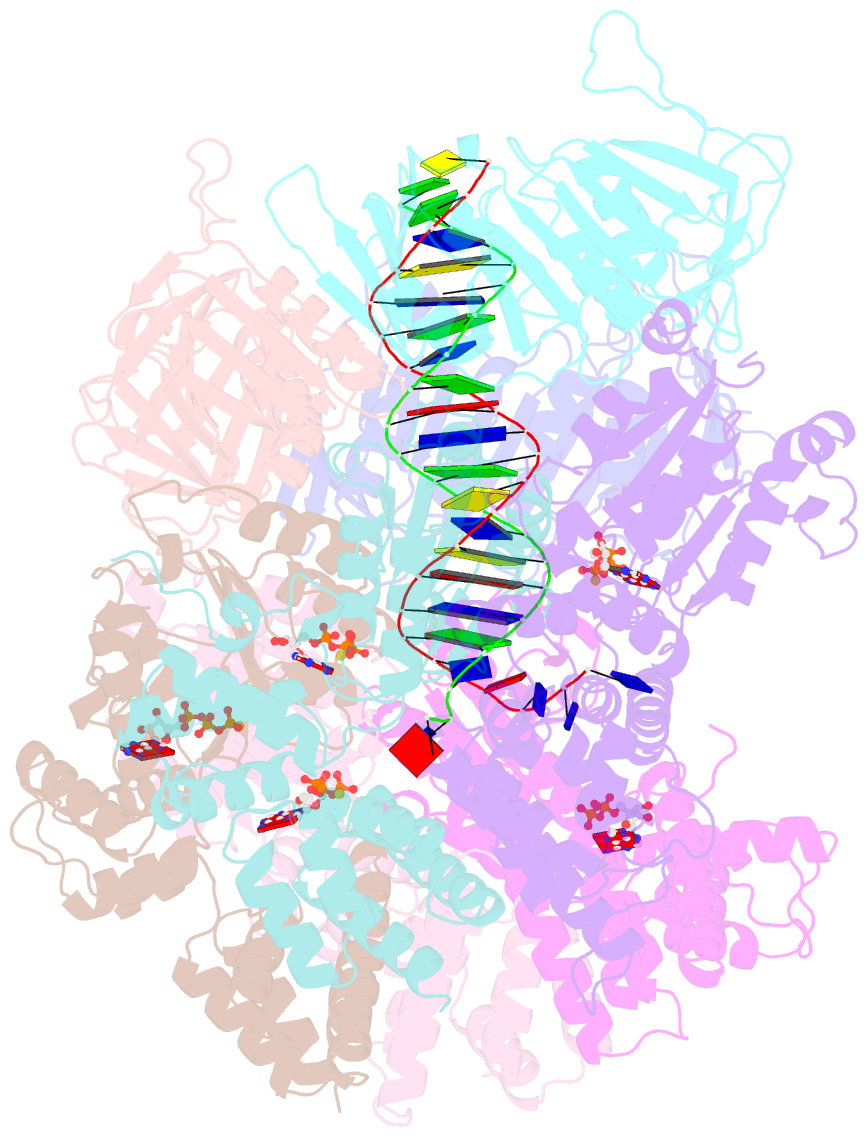
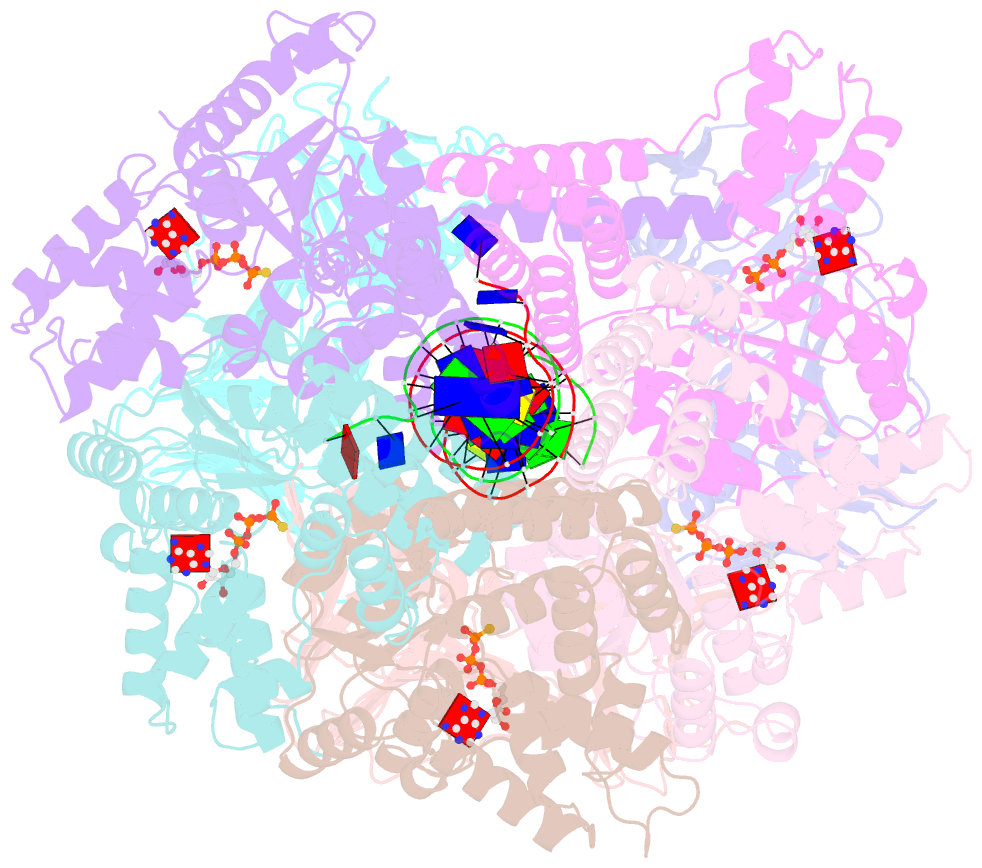
Summary information and primary citation
- PDB-id
- 7tfi; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- cryo-EM (3.41 Å)
- Summary
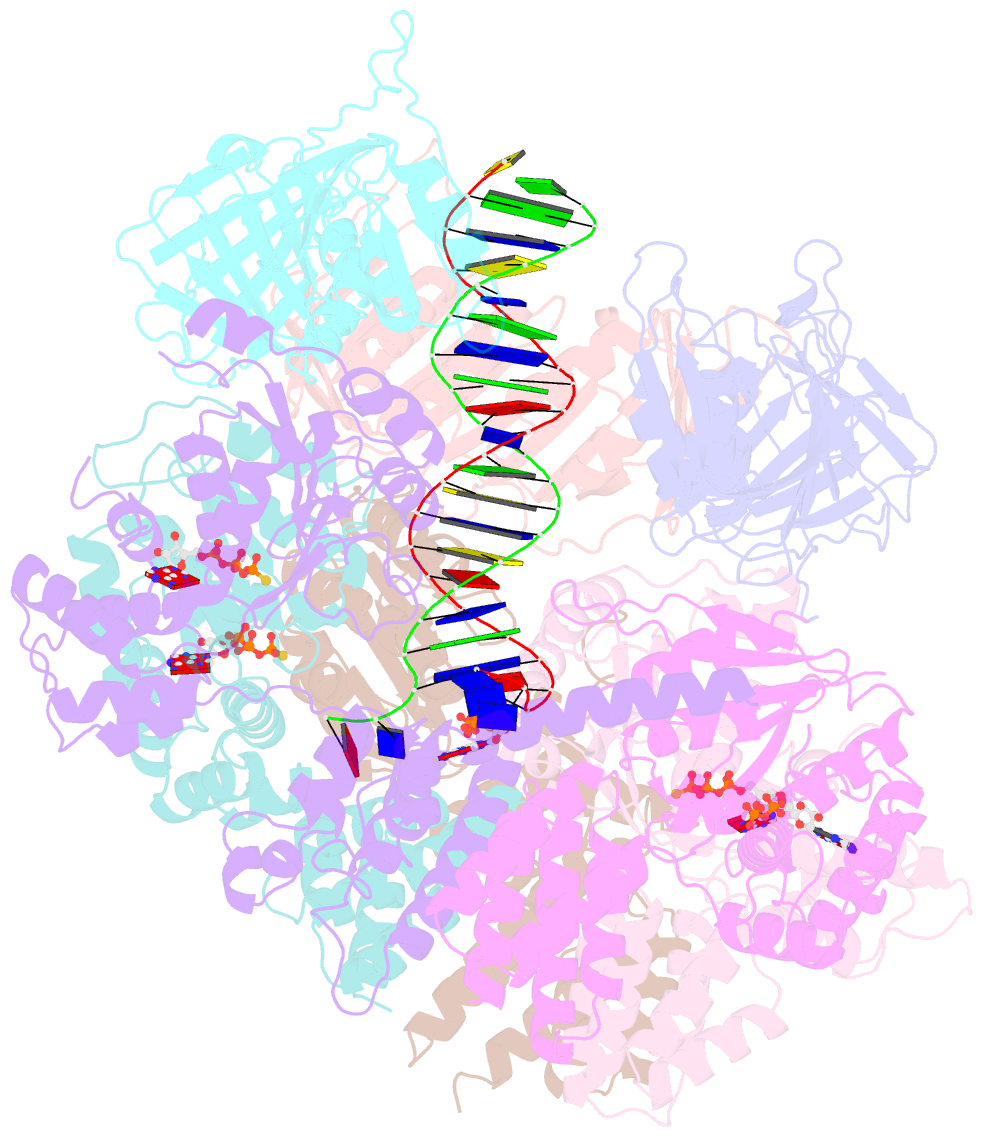
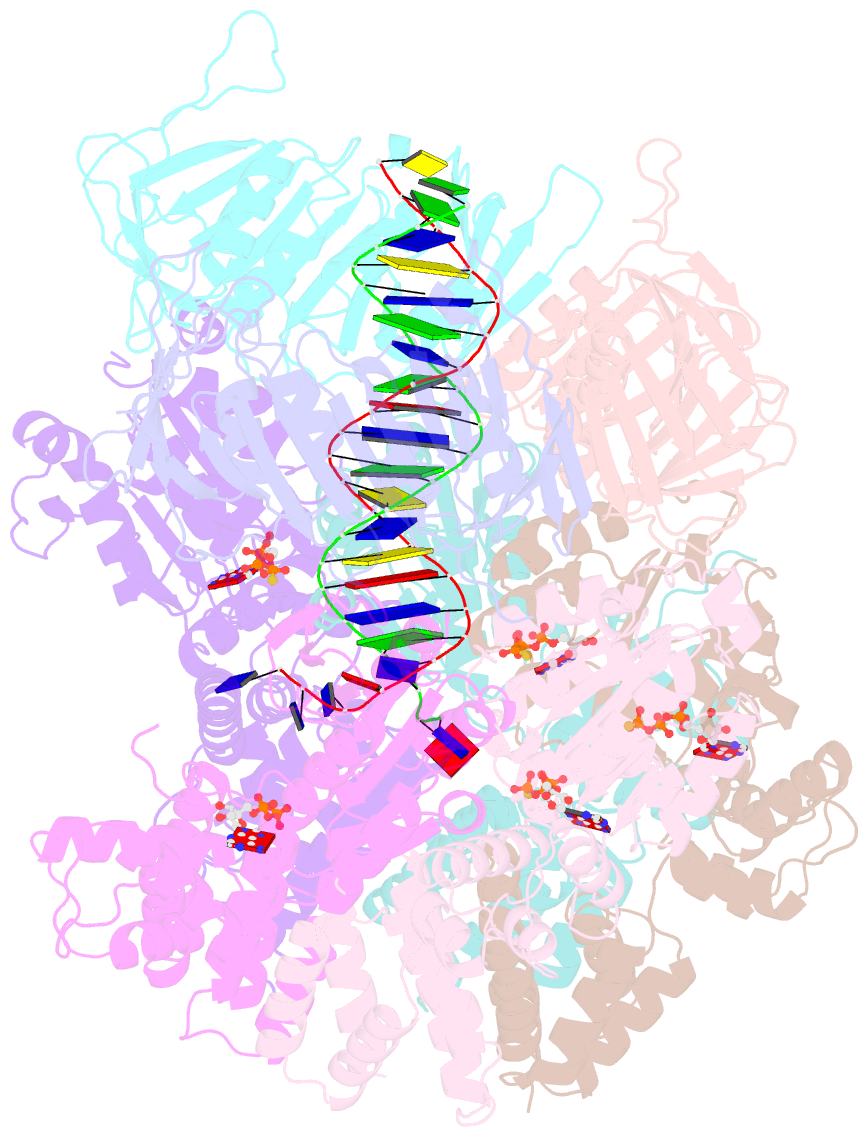
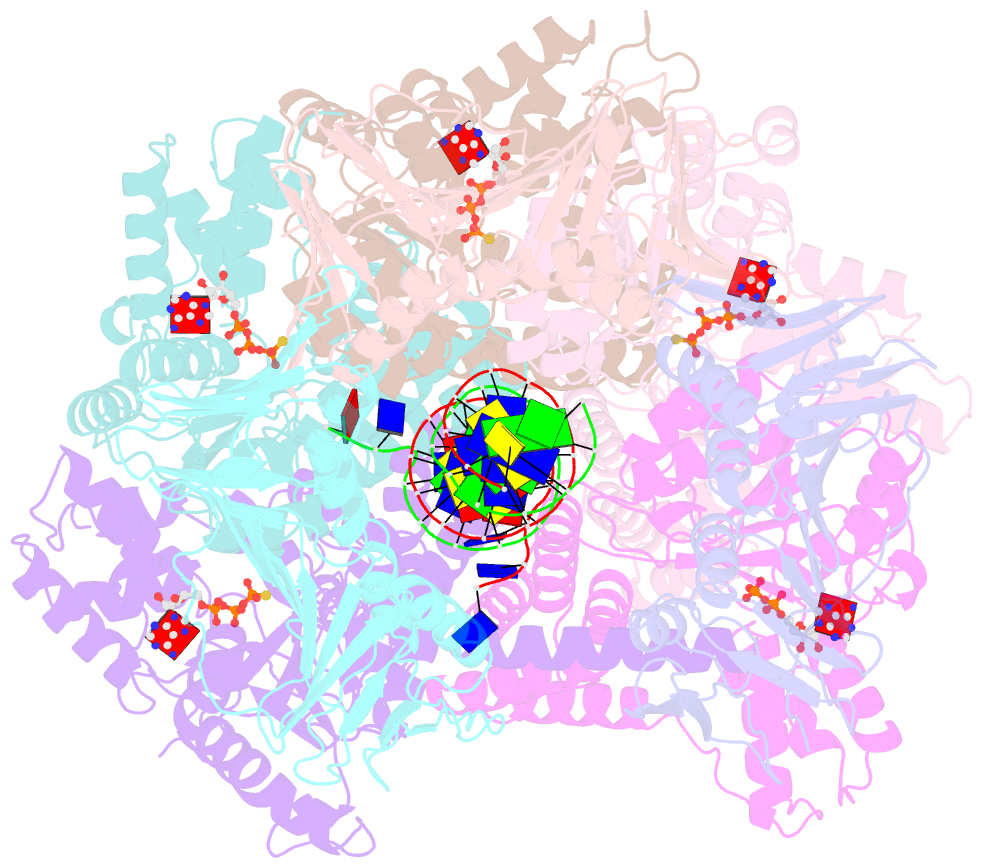
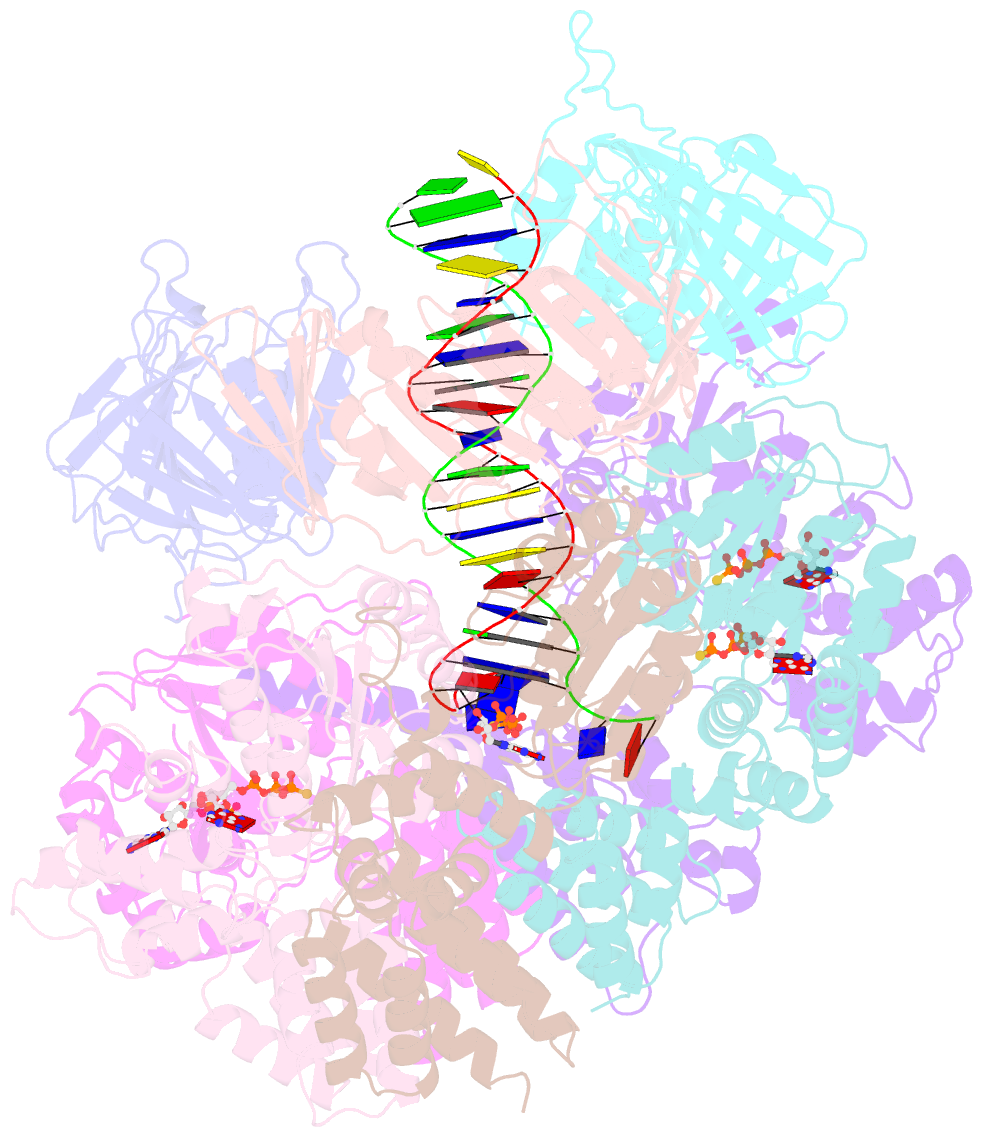
- Atomic model of the s. cerevisiae clamp-clamp loader complex pcna-rfc bound to DNA with an open clamp
- Reference
- Zheng F, Georgescu R, Yao NY, Li H, O'Donnell ME (2022): "Cryo-EM structures reveal that RFC recognizes both the 3'- and 5'-DNA ends to load PCNA onto gaps for DNA repair." Elife, 11. doi: 10.7554/eLife.77469.
- Abstract
- RFC uses ATP to assemble PCNA onto primed sites for replicative DNA polymerases δ and ε. The RFC pentamer forms a central chamber that binds 3' ss/ds DNA junctions to load PCNA onto DNA during replication. We show here five structures that identify a second DNA binding site in RFC that binds a 5' duplex. This 5' DNA site is located between the N-terminal BRCT domain and AAA+ module of the large Rfc1 subunit. Our structures reveal ideal binding to a 7-nt gap, which includes 2 bp unwound by the clamp loader. Biochemical studies show enhanced binding to 5 and 10 nt gaps, consistent with the structural results. Because both 3' and 5' ends are present at a ssDNA gap, we propose that the 5' site facilitates RFC's PCNA loading activity at a DNA damage-induced gap to recruit gap-filling polymerases. These findings are consistent with genetic studies showing that base excision repair of gaps greater than 1 base requires PCNA and involves the 5' DNA binding domain of Rfc1. We further observe that a 5' end facilitates PCNA loading at an RPA coated 30-nt gap, suggesting a potential role of the RFC 5'-DNA site in lagging strand DNA synthesis.