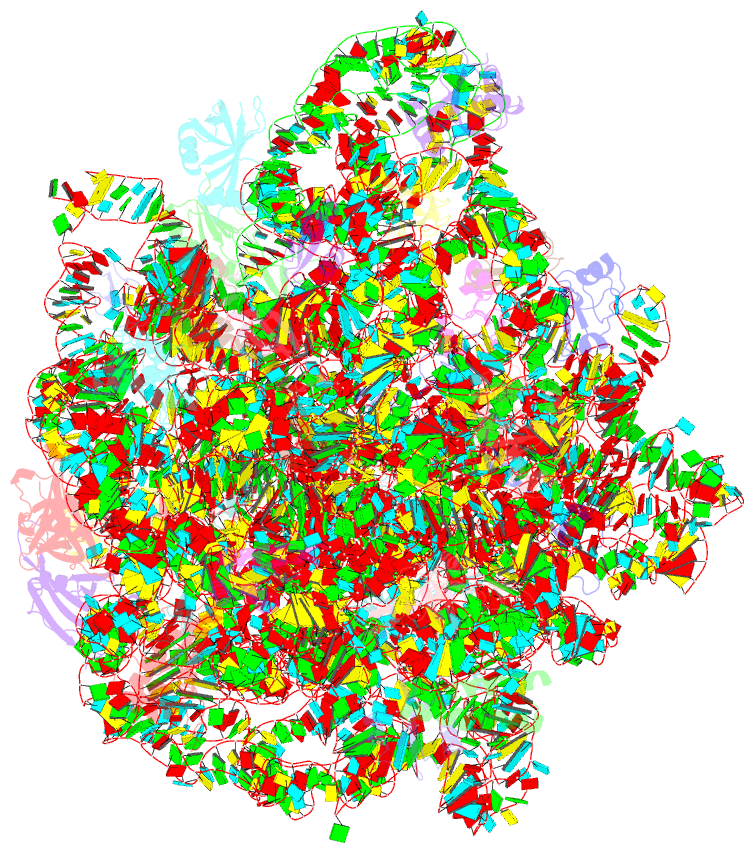
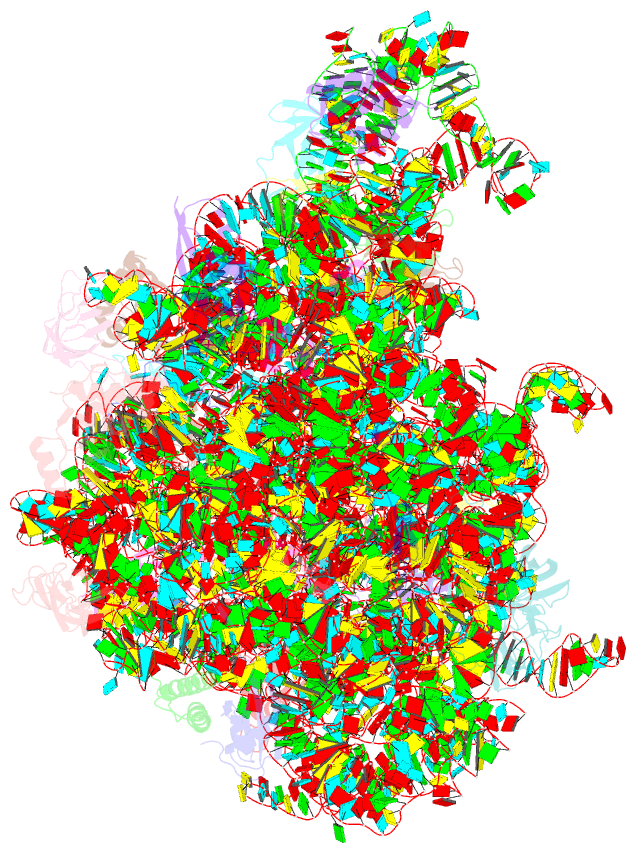
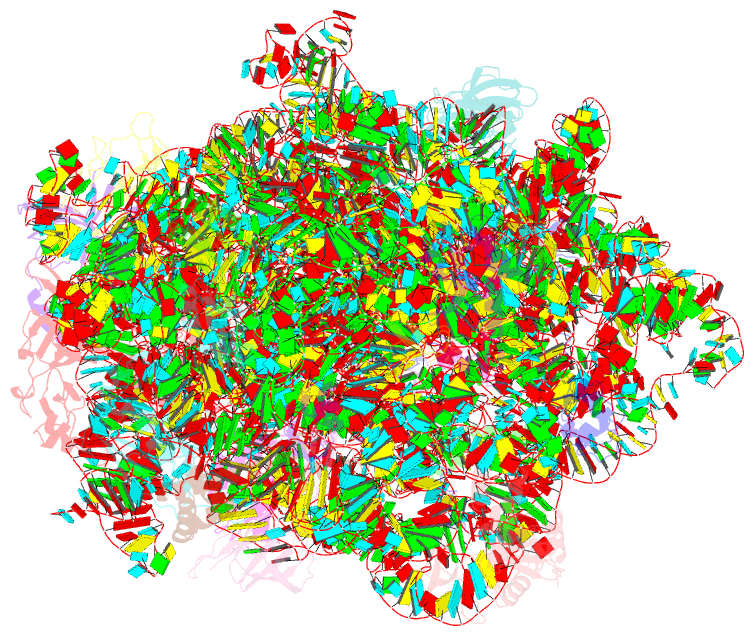
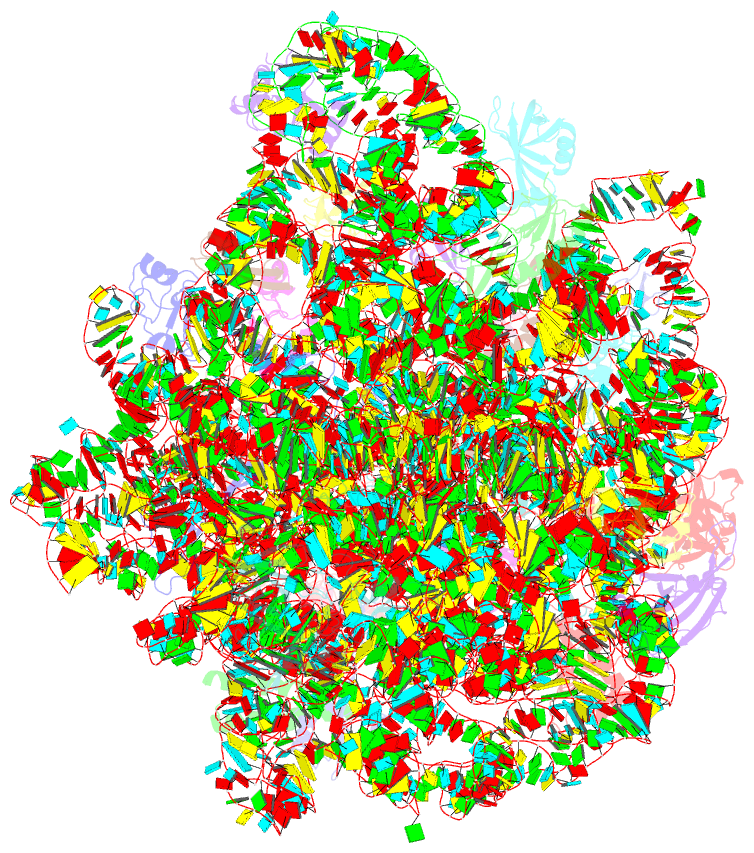
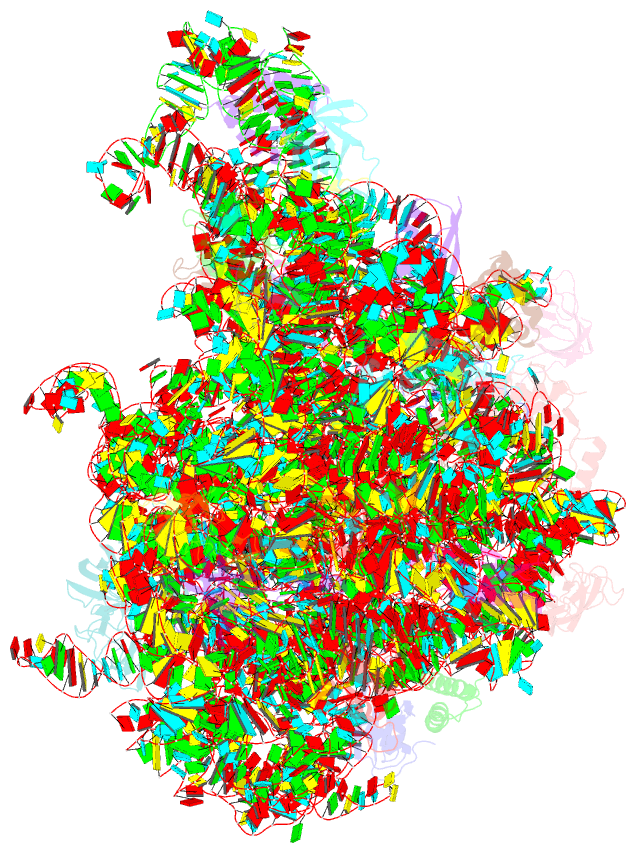
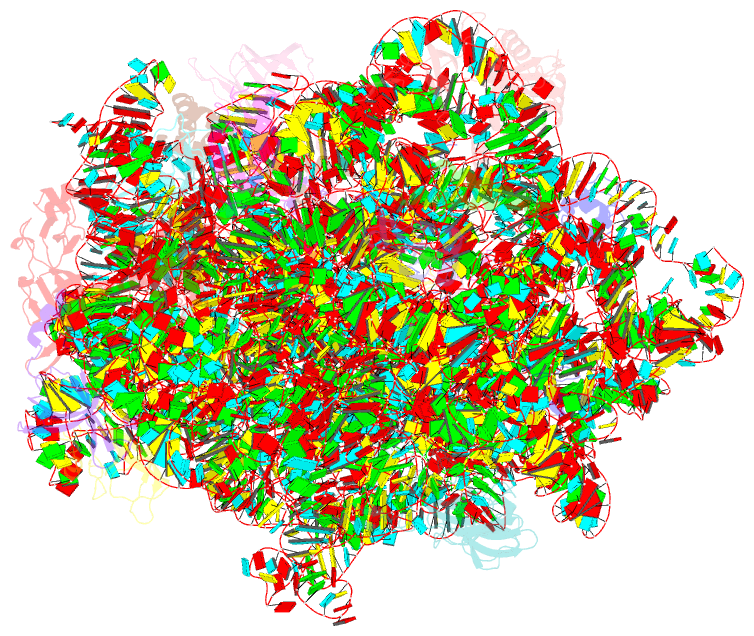
Summary information and primary citation
- PDB-id
- 7ttw; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ribosome
- Method
- cryo-EM (2.9 Å)
- Summary
- 50s ribosomal subunit from staphylococcus aureus containing double mutation in ul3 imparting linezolid resistance
- Reference
- Perlaza-Jimenez L, Tan KS, Piper SJ, Johnson RM, Bamert RS, Stubenrauch CJ, Wright A, Lupton D, Lithgow T, Belousoff MJ (2022): "A Structurally Characterized Staphylococcus aureus Evolutionary Escape Route from Treatment with the Antibiotic Linezolid." Microbiol Spectr, 10, e0058322. doi: 10.1128/spectrum.00583-22.
- Abstract
- Methicillin-resistant Staphylococcus aureus (MRSA) is a bacterial pathogen that presents great health concerns. Treatment requires the use of last-line antibiotics, such as members of the oxazolidinone family, of which linezolid is the first member to see regular use in the clinic. Here, we report a short time scale selection experiment in which strains of MRSA were subjected to linezolid treatment. Clonal isolates which had evolved a linezolid-resistant phenotype were characterized by whole-genome sequencing. Linezolid-resistant mutants were identified which had accumulated mutations in the ribosomal protein uL3. Multiple clones which had two mutations in uL3 exhibited resistance to linezolid, 2-fold higher than the clinical breakpoint. Ribosomes from this strain were isolated and subjected to single-particle cryo-electron microscopic analysis and compared to the ribosomes from the parent strain. We found that the mutations in uL3 lead to a rearrangement of a loop that makes contact with Helix 90, propagating a structural change over 15 Å away. This distal change swings nucleotide U2504 into the binding site of the antibiotic, causing linezolid resistance.