Summary information and primary citation
- PDB-id
- 7u22; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- antibiotic
- Method
- X-ray (3.87 Å)
- Summary
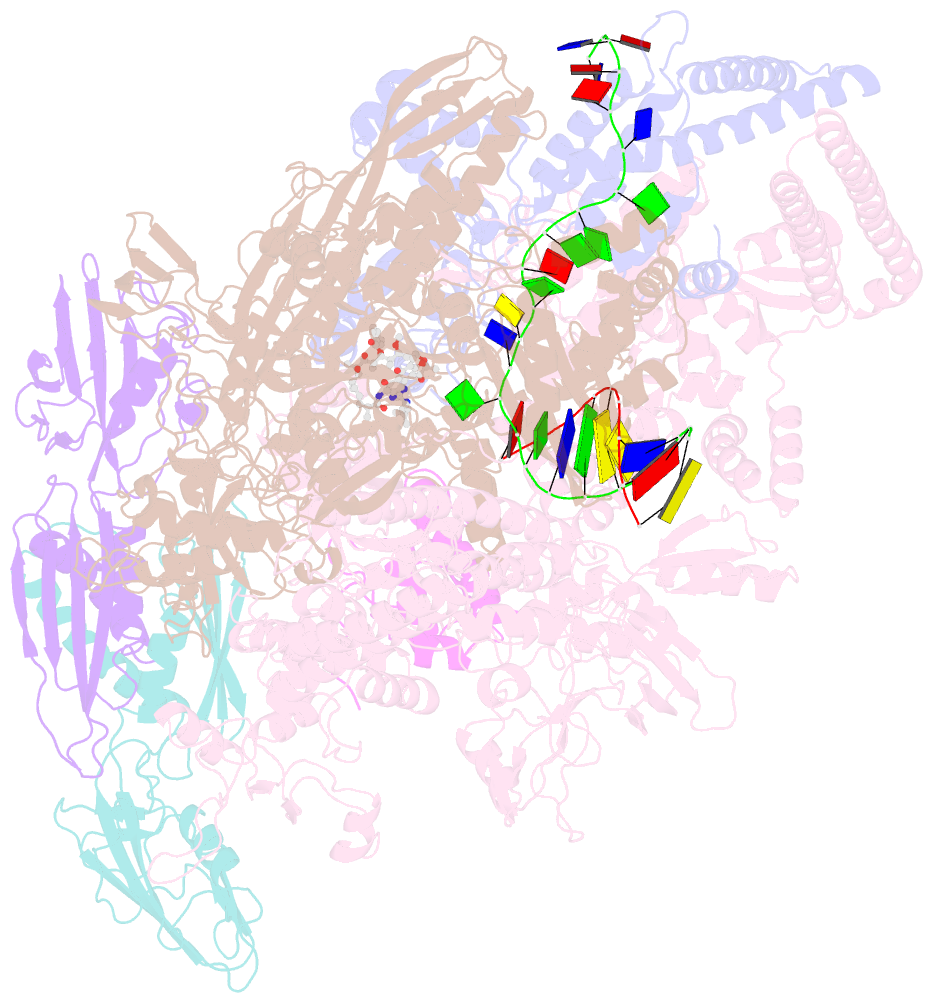
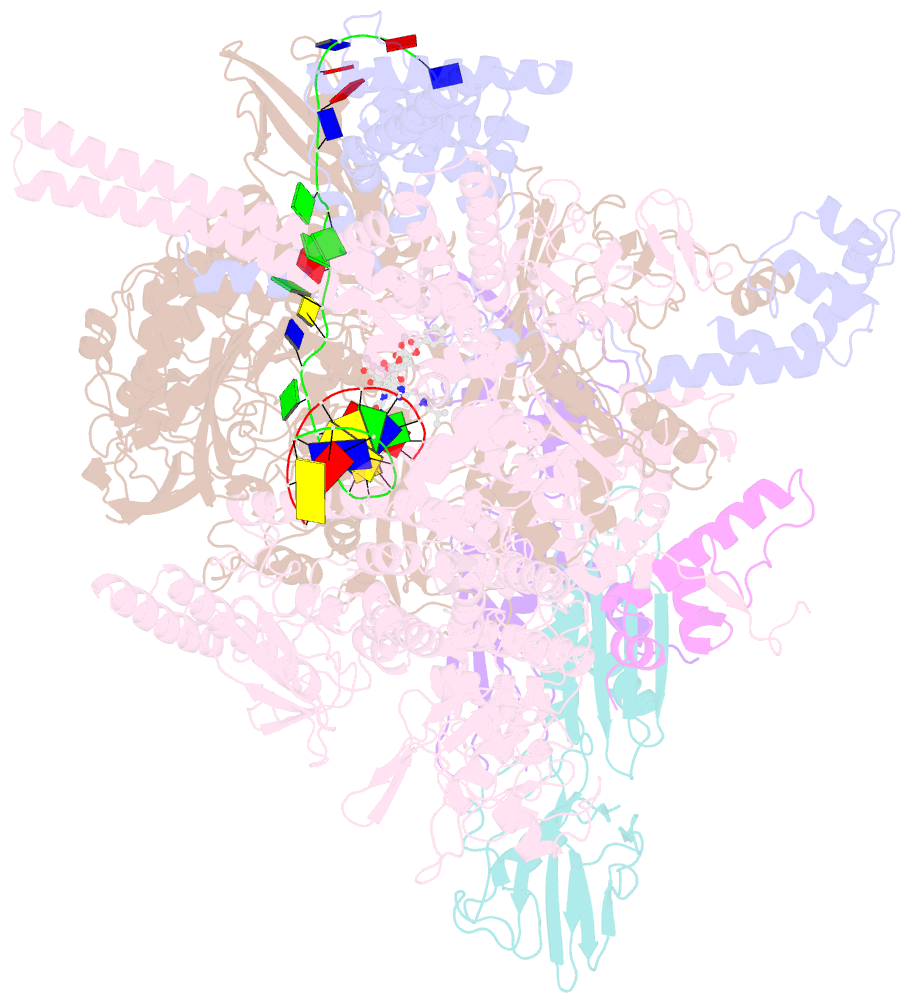
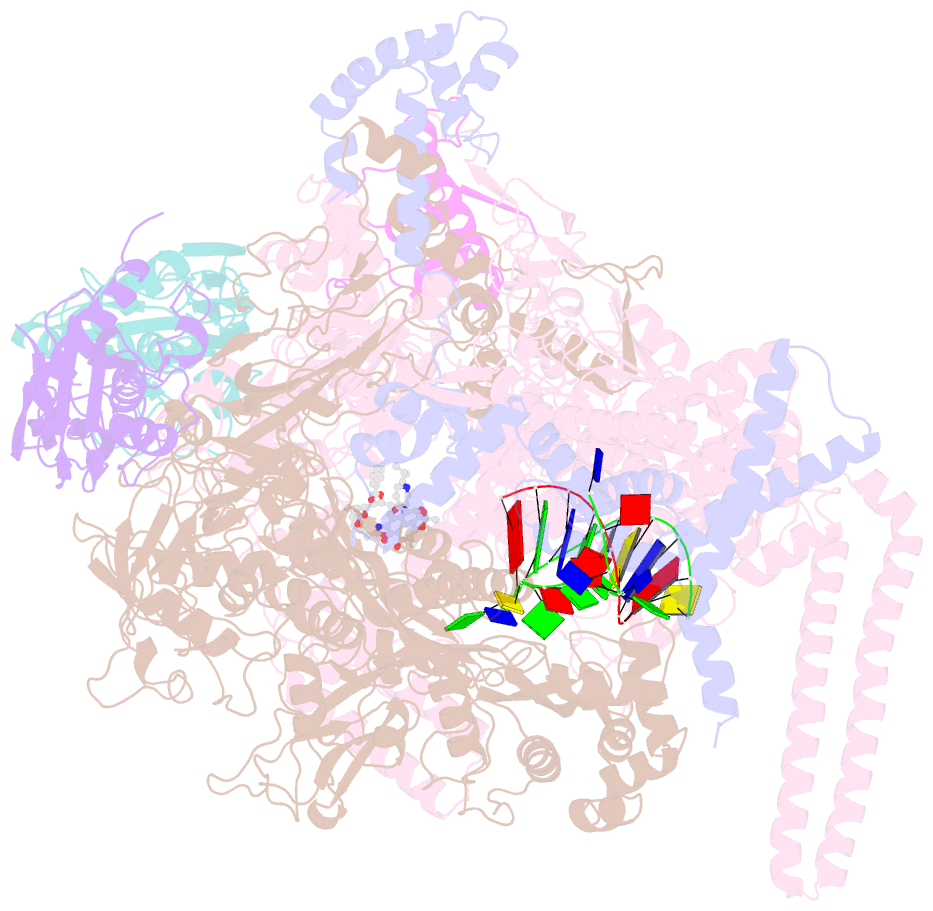
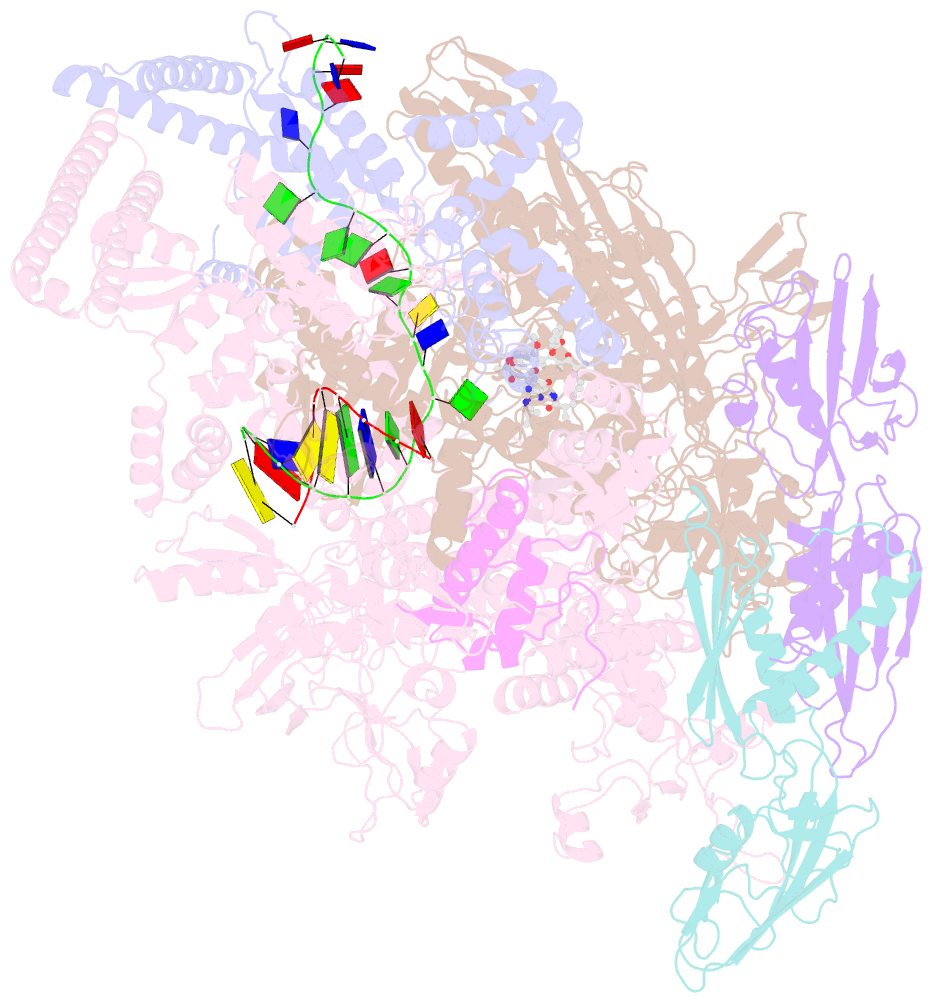
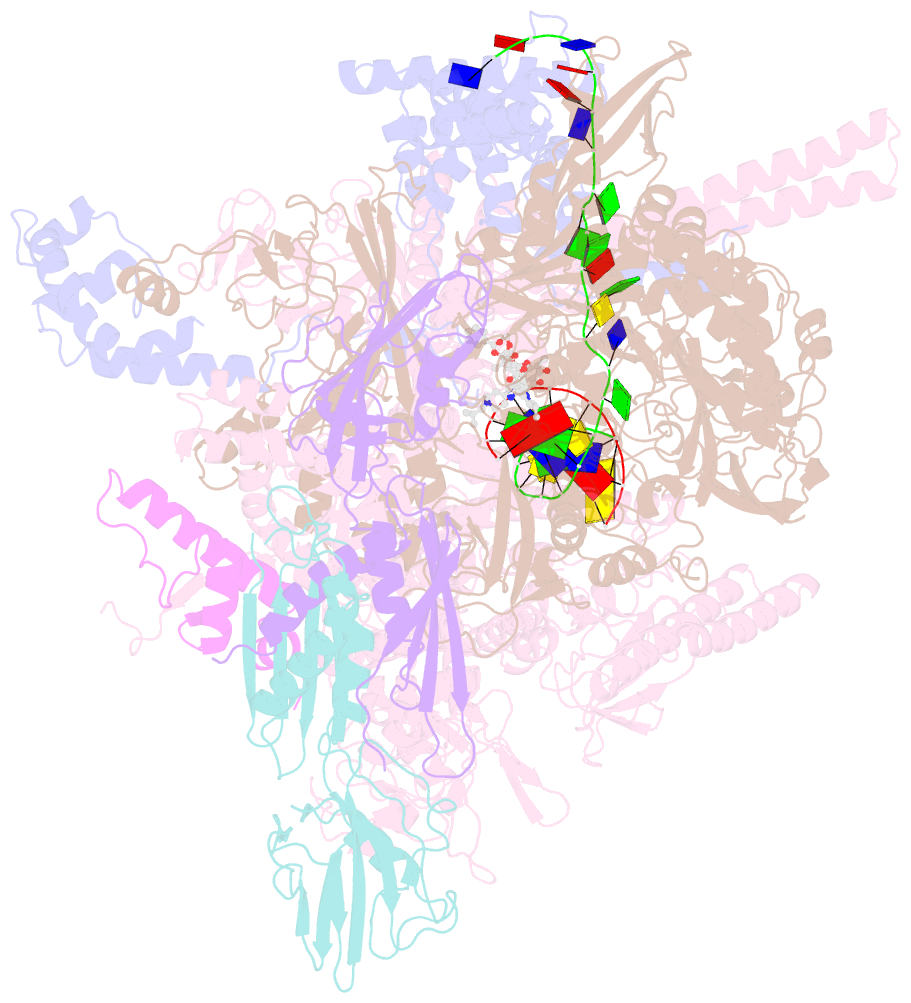
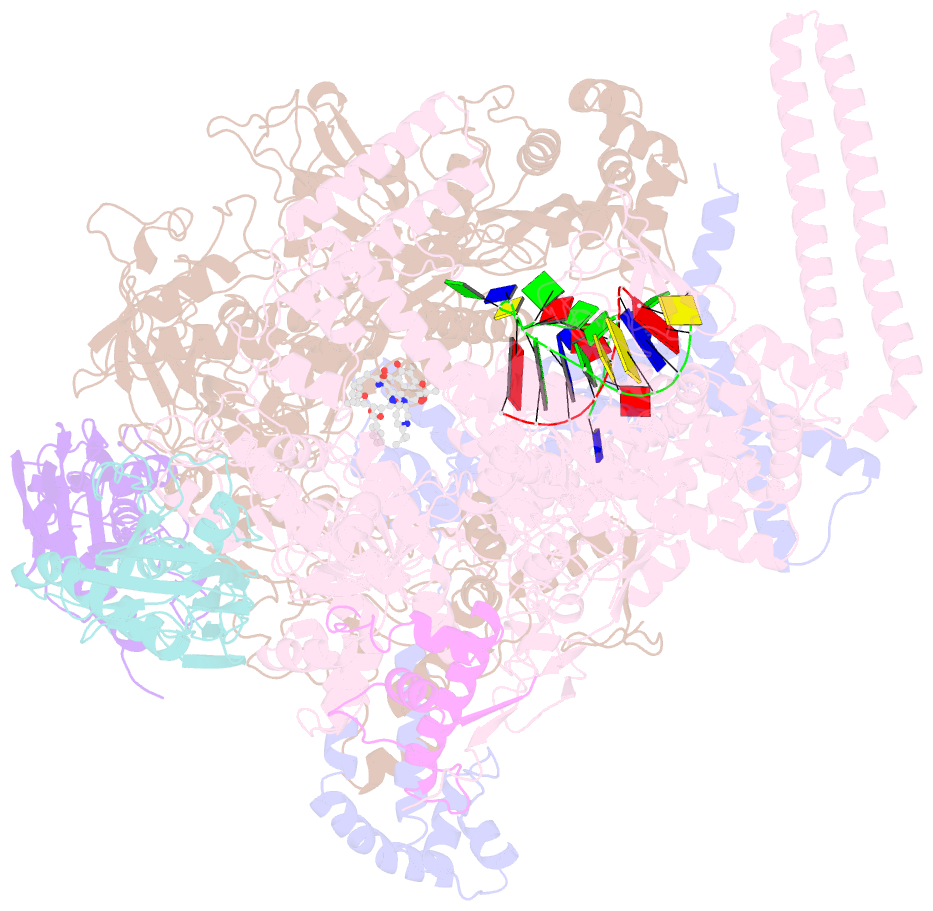
- Mycobacterium tuberculosis RNA polymerase sigma a holoenzyme open promoter complex containing umn-7
- Reference
- Lan T, Ganapathy US, Sharma S, Ahn YM, Zimmerman M, Molodtsov V, Hegde P, Gengenbacher M, Ebright RH, Dartois V, Freundlich JS, Dick T, Aldrich CC (2022): "Redesign of Rifamycin Antibiotics to Overcome ADP-Ribosylation-Mediated Resistance." Angew.Chem.Int.Ed.Engl., 61, e202211498. doi: 10.1002/anie.202211498.
- Abstract
- Rifamycin antibiotics are a valuable class of antimicrobials for treating infections by mycobacteria and other persistent bacteria owing to their potent bactericidal activity against replicating and non-replicating pathogens. However, the clinical utility of rifamycins against Mycobacterium abscessus is seriously compromised by a novel resistance mechanism, namely, rifamycin inactivation by ADP-ribosylation. Using a structure-based approach, we rationally redesign rifamycins through strategic modification of the ansa-chain to block ADP-ribosylation while preserving on-target activity. Validated by a combination of biochemical, structural, and microbiological studies, the most potent analogs overcome ADP-ribosylation, restored their intrinsic low nanomolar activity and demonstrated significant in vivo antibacterial efficacy. Further optimization by tuning drug disposition properties afforded a preclinical candidate with remarkable potency and an outstanding pharmacokinetic profile.