Summary information and primary citation
- PDB-id
- 7u32; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein-DNA
- Method
- cryo-EM (3.46 Å)
- Summary
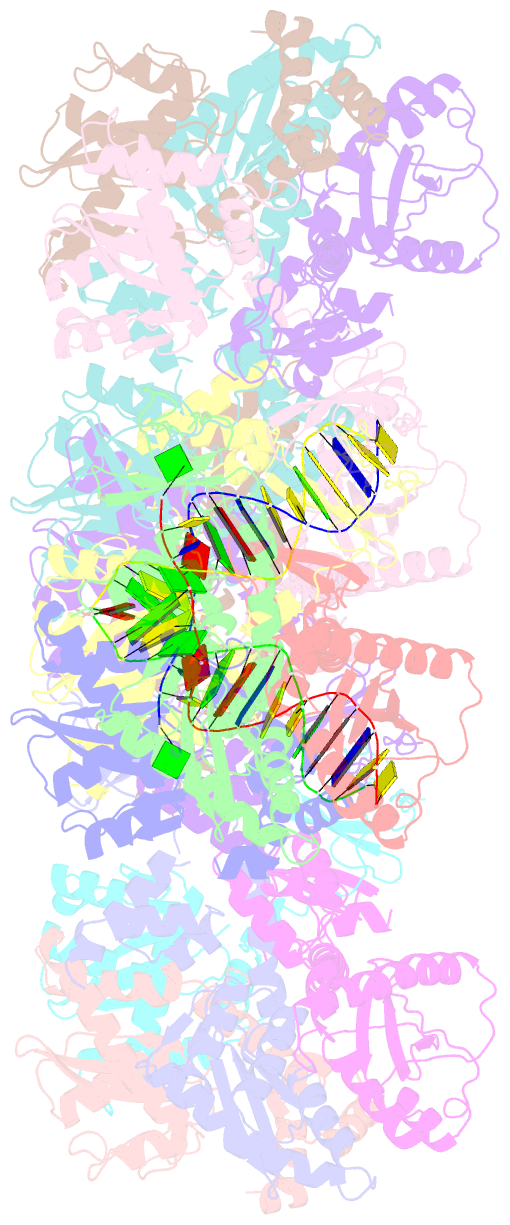
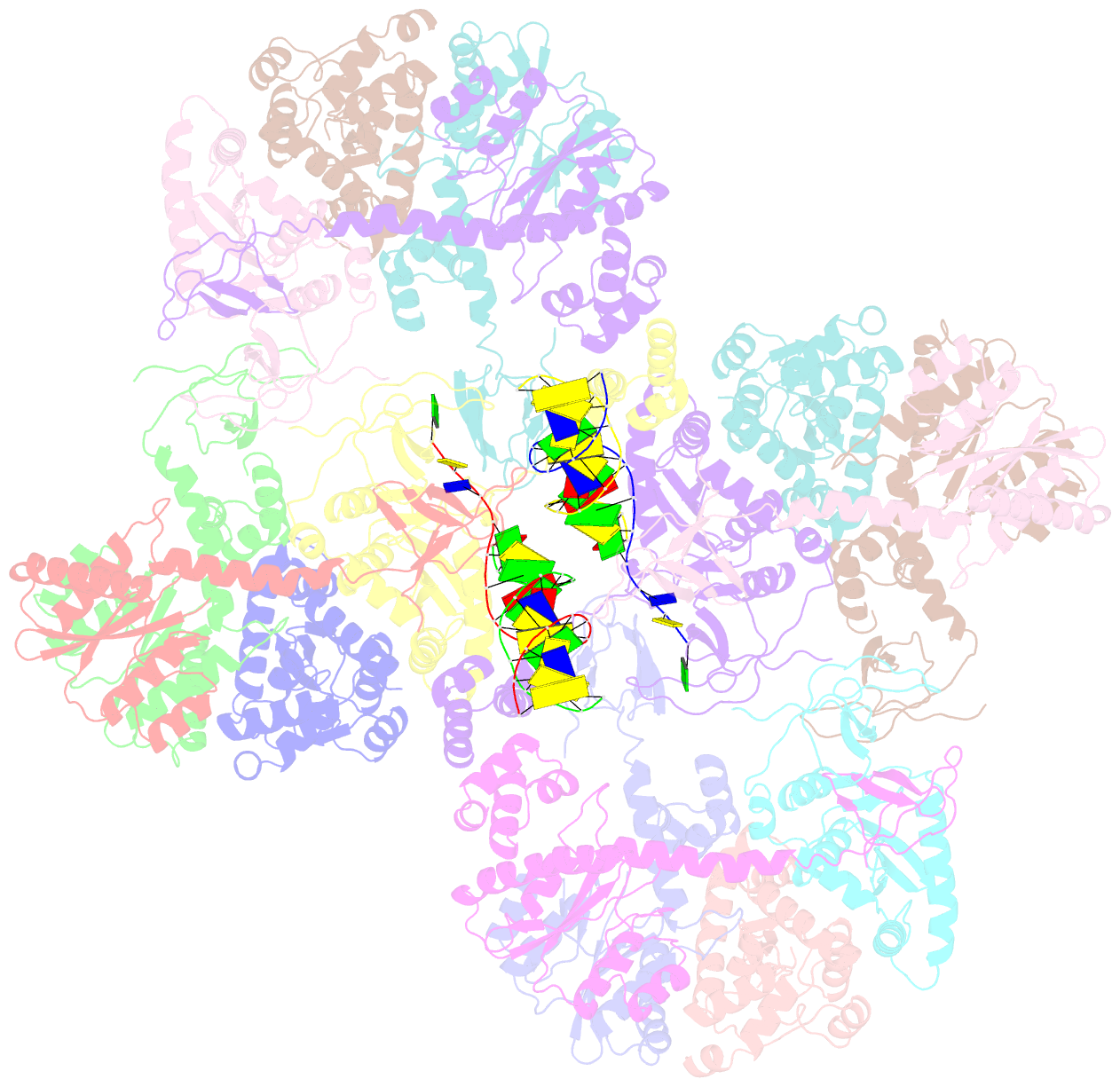
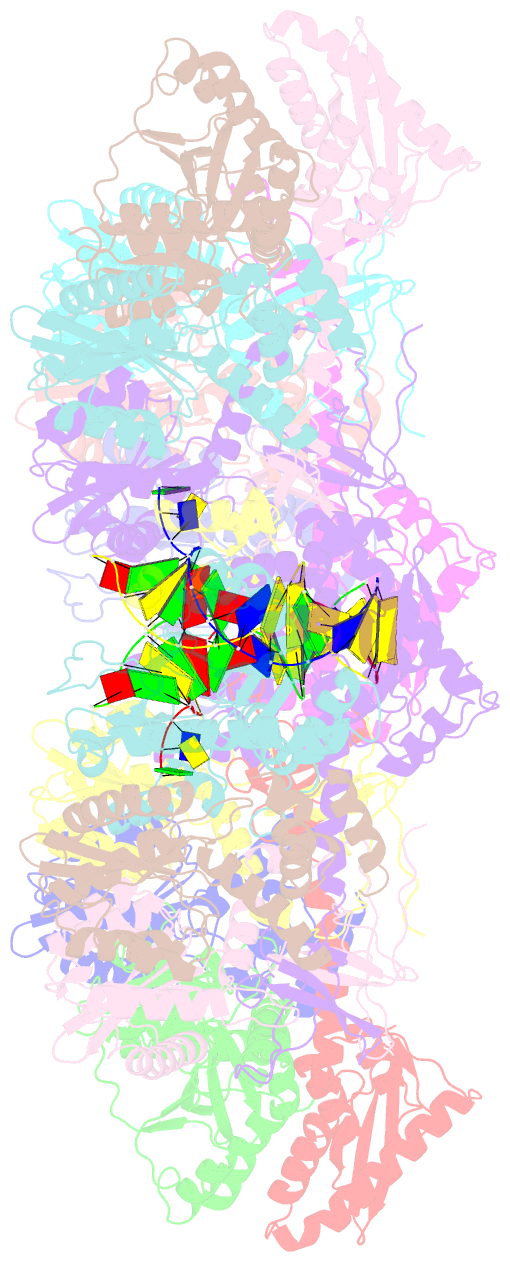
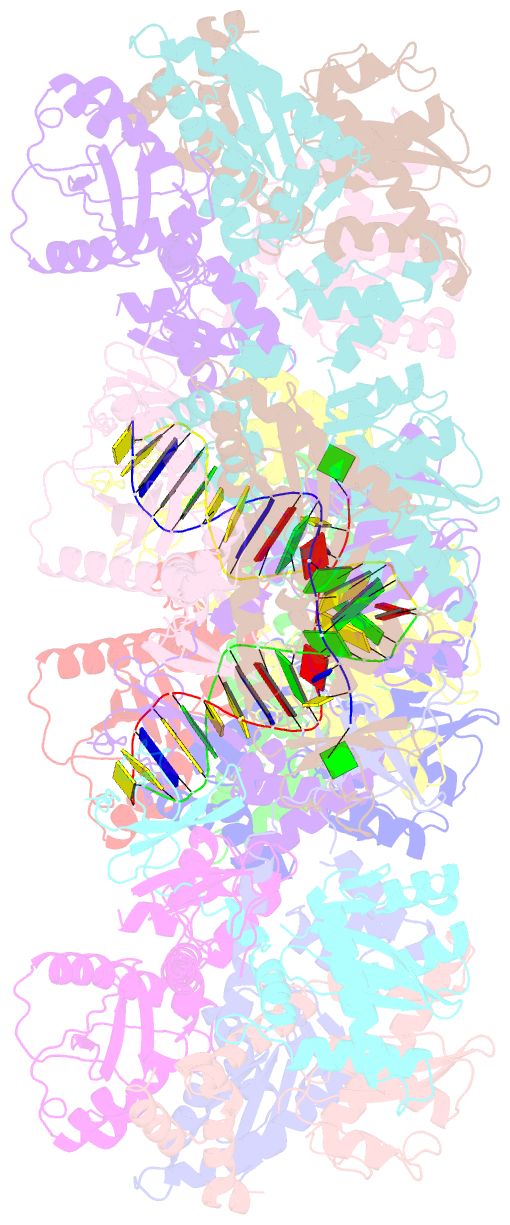
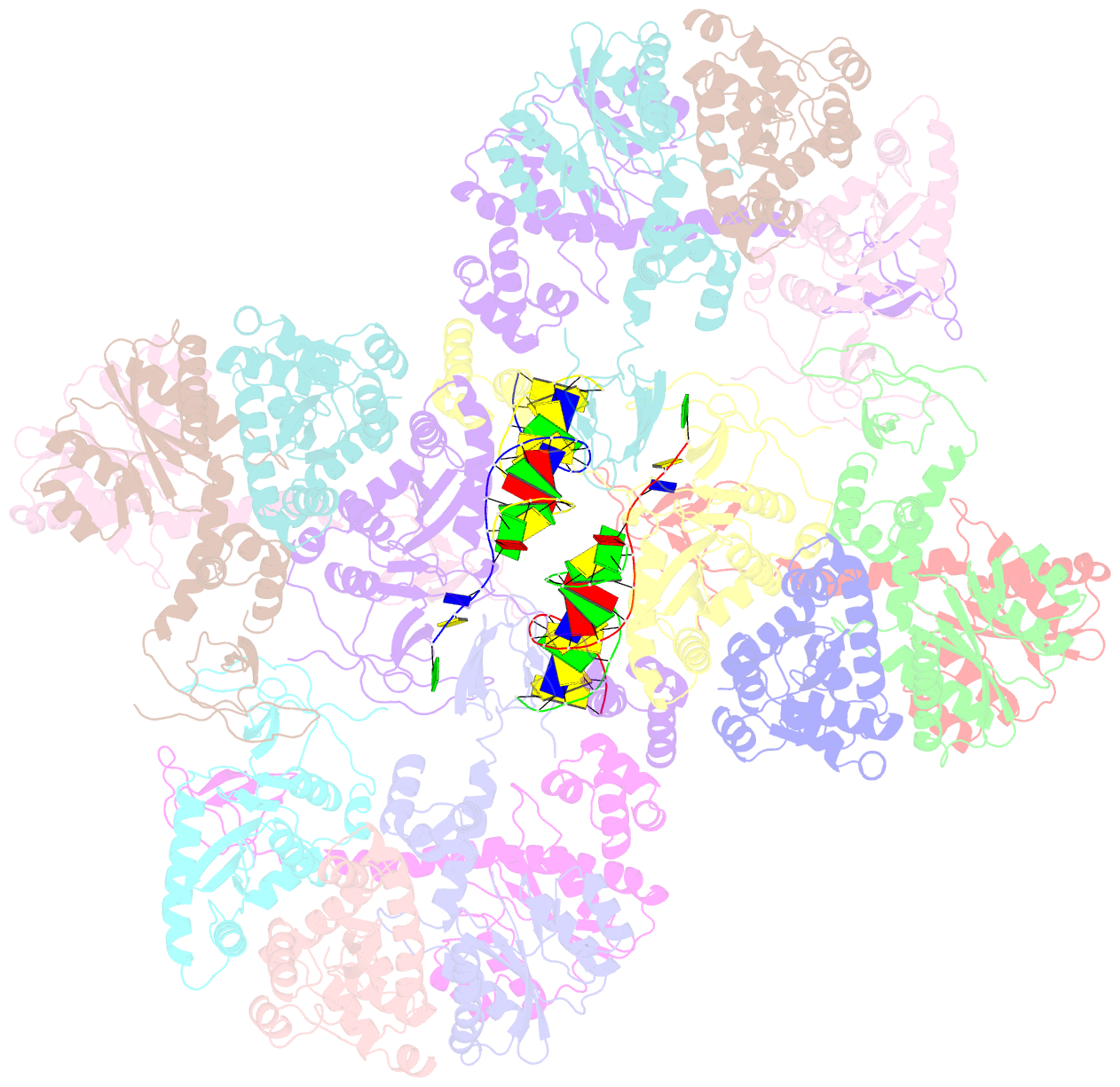
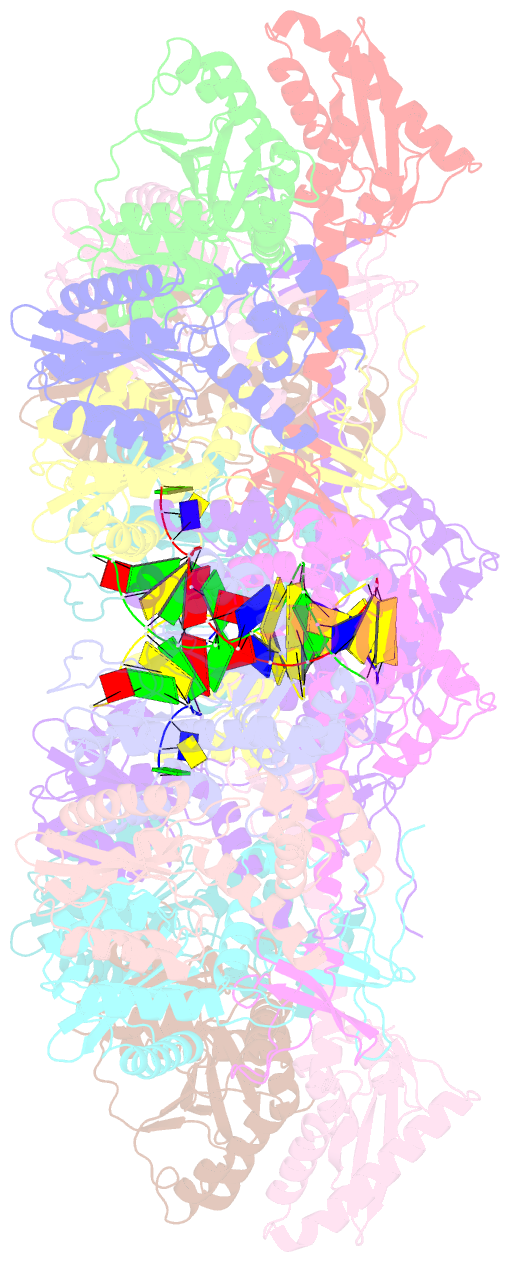
- Mvv cleaved synaptic complex (csc) intasome at 3.4 Å resolution
- Reference
- Ballandras-Colas A, Chivukula V, Gruszka DT, Shan Z, Singh PK, Pye VE, McLean RK, Bedwell GJ, Li W, Nans A, Cook NJ, Fadel HJ, Poeschla EM, Griffiths DJ, Vargas J, Taylor IA, Lyumkis D, Yardimci H, Engelman AN, Cherepanov P (2022): "Multivalent interactions essential for lentiviral integrase function." Nat Commun, 13, 2416. doi: 10.1038/s41467-022-29928-8.
- Abstract
- A multimer of retroviral integrase (IN) synapses viral DNA ends within a stable intasome nucleoprotein complex for integration into a host cell genome. Reconstitution of the intasome from the maedi-visna virus (MVV), an ovine lentivirus, revealed a large assembly containing sixteen IN subunits1. Herein, we report cryo-EM structures of the lentiviral intasome prior to engagement of target DNA and following strand transfer, refined at 3.4 and 3.5 Å resolution, respectively. The structures elucidate details of the protein-protein and protein-DNA interfaces involved in lentiviral intasome formation. We show that the homomeric interfaces involved in IN hexadecamer formation and the α-helical configuration of the linker connecting the C-terminal and catalytic core domains are critical for MVV IN strand transfer activity in vitro and for virus infectivity. Single-molecule microscopy in conjunction with photobleaching reveals that the MVV intasome can bind a variable number, up to sixteen molecules, of the lentivirus-specific host factor LEDGF/p75. Concordantly, ablation of endogenous LEDGF/p75 results in gross redistribution of MVV integration sites in human and ovine cells. Our data confirm the importance of the expanded architecture observed in cryo-EM studies of lentiviral intasomes and suggest that this organization underlies multivalent interactions with chromatin for integration targeting to active genes.