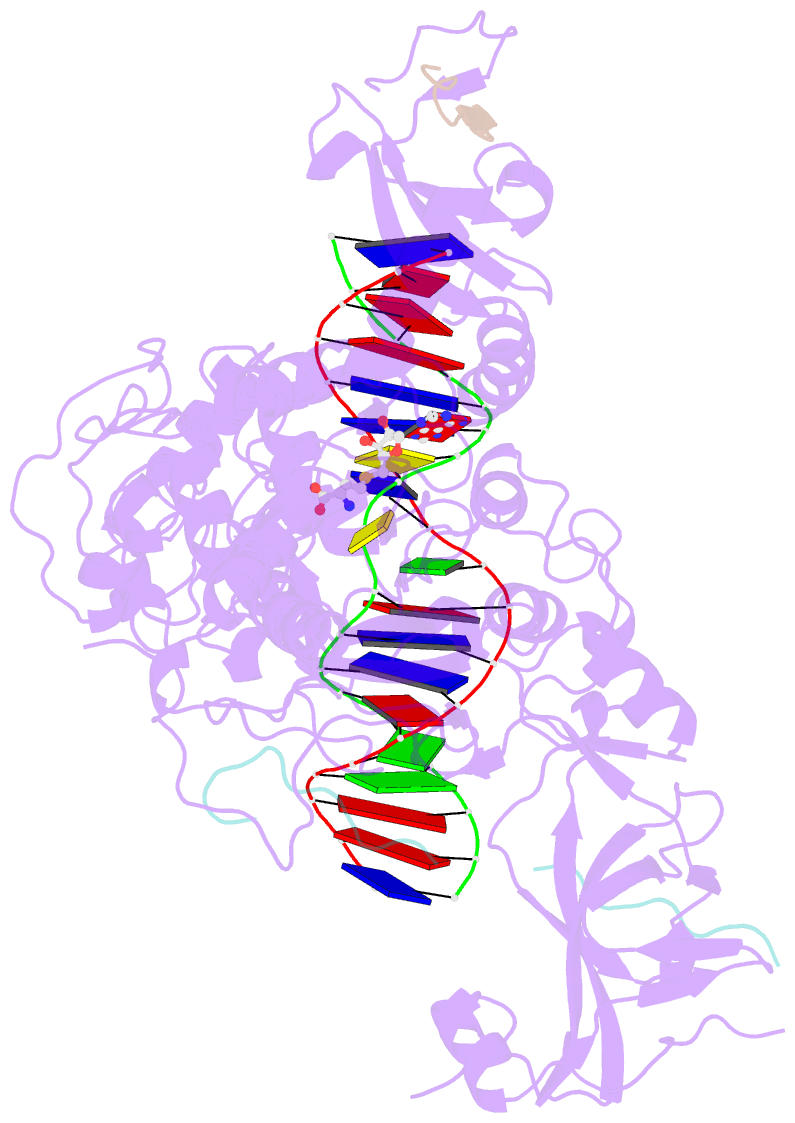
Summary information and primary citation
- PDB-id
- 7ubu; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.39 Å)
- Summary
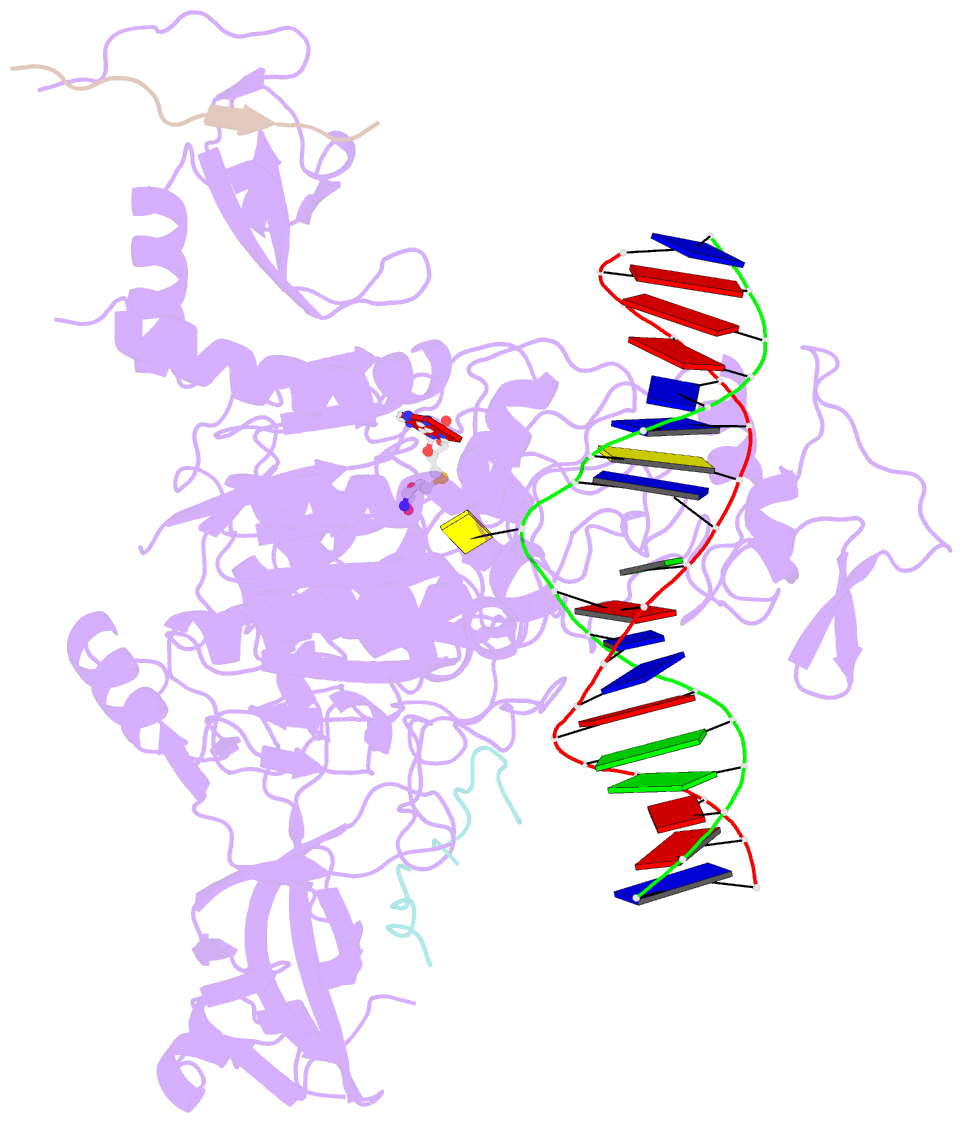
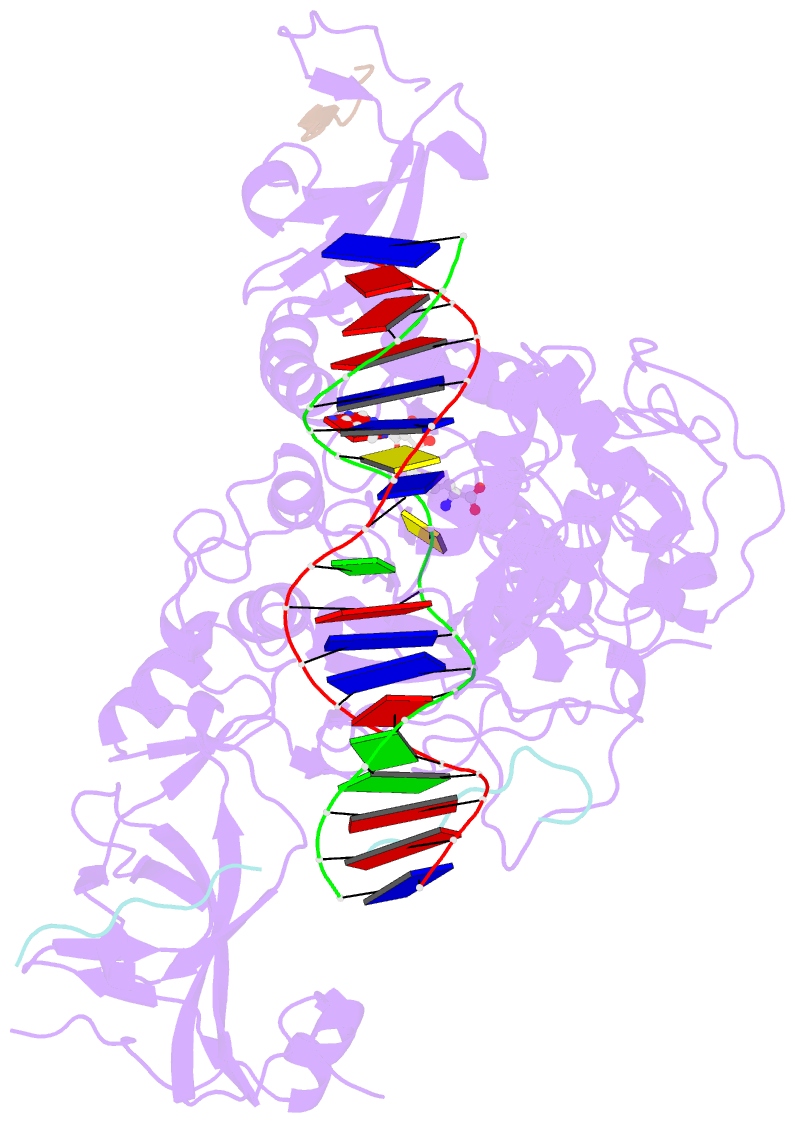
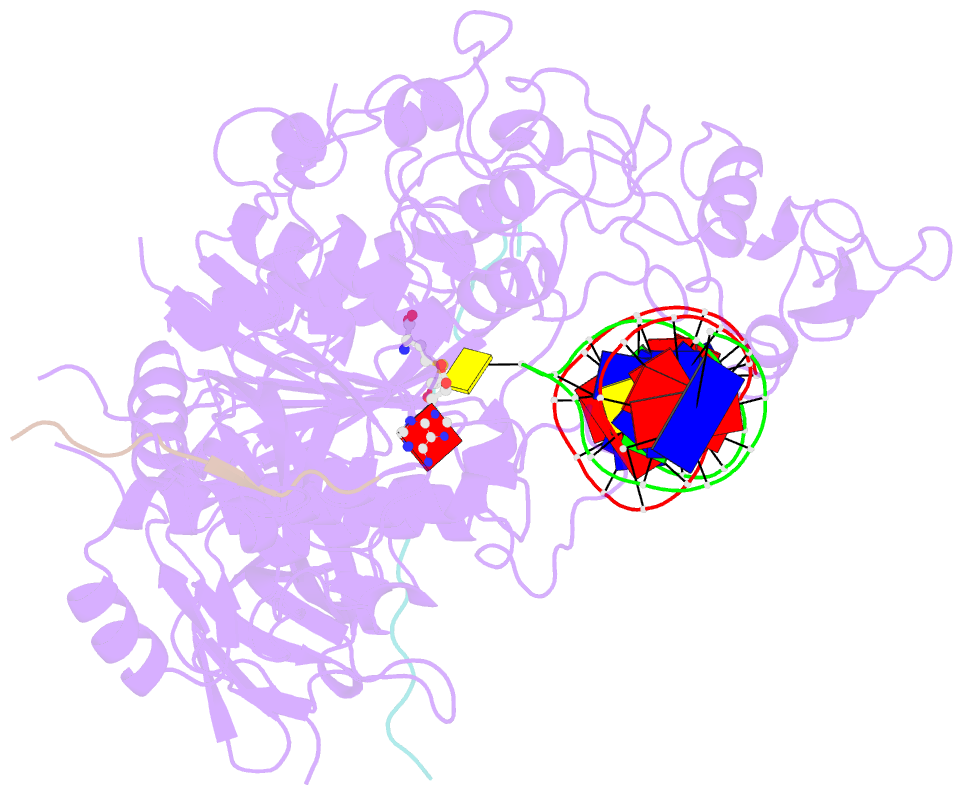
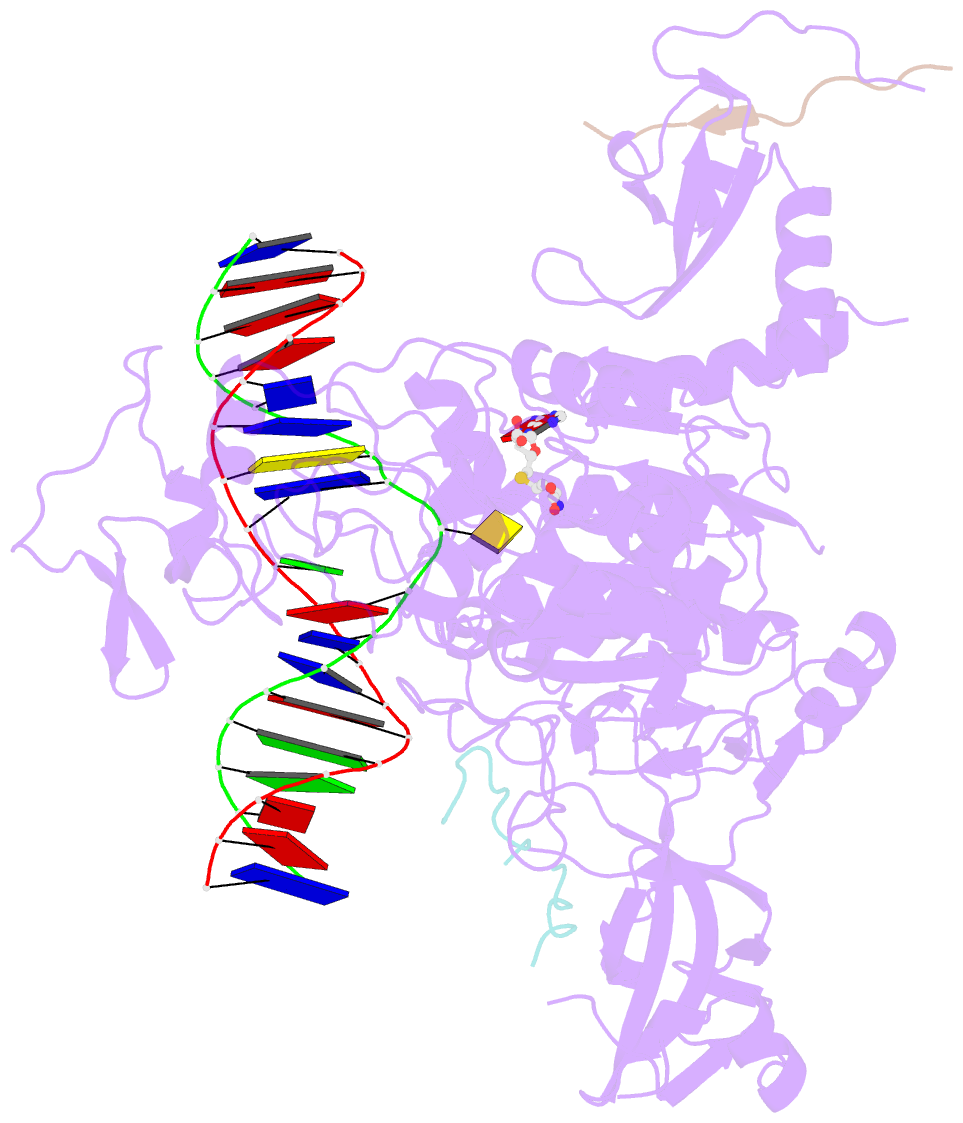
- Crystal structure of zmet2 in complex with hemimethylated cag DNA and a histone h3kc9me2 peptide
- Reference
- Fang J, Jiang J, Leichter SM, Liu J, Biswal M, Khudaverdyan N, Zhong X, Song J (2022): "Mechanistic basis for maintenance of CHG DNA methylation in plants." Nat Commun, 13, 3877. doi: 10.1038/s41467-022-31627-3.
- Abstract
- DNA methylation is an evolutionarily conserved epigenetic mechanism essential for transposon silencing and heterochromatin assembly. In plants, DNA methylation widely occurs in the CG, CHG, and CHH (H = A, C, or T) contexts, with the maintenance of CHG methylation mediated by CMT3 chromomethylase. However, how CMT3 interacts with the chromatin environment for faithful maintenance of CHG methylation is unclear. Here we report structure-function characterization of the H3K9me2-directed maintenance of CHG methylation by CMT3 and its Zea mays ortholog ZMET2. Base-specific interactions and DNA deformation coordinately underpin the substrate specificity of CMT3 and ZMET2, while a bivalent readout of H3K9me2 and H3K18 allosterically stimulates substrate binding. Disruption of the interaction with DNA or H3K9me2/H3K18 led to loss of CMT3/ZMET2 activity in vitro and impairment of genome-wide CHG methylation in vivo. Together, our study uncovers how the intricate interplay of CMT3, repressive histone marks, and DNA sequence mediates heterochromatic CHG methylation.