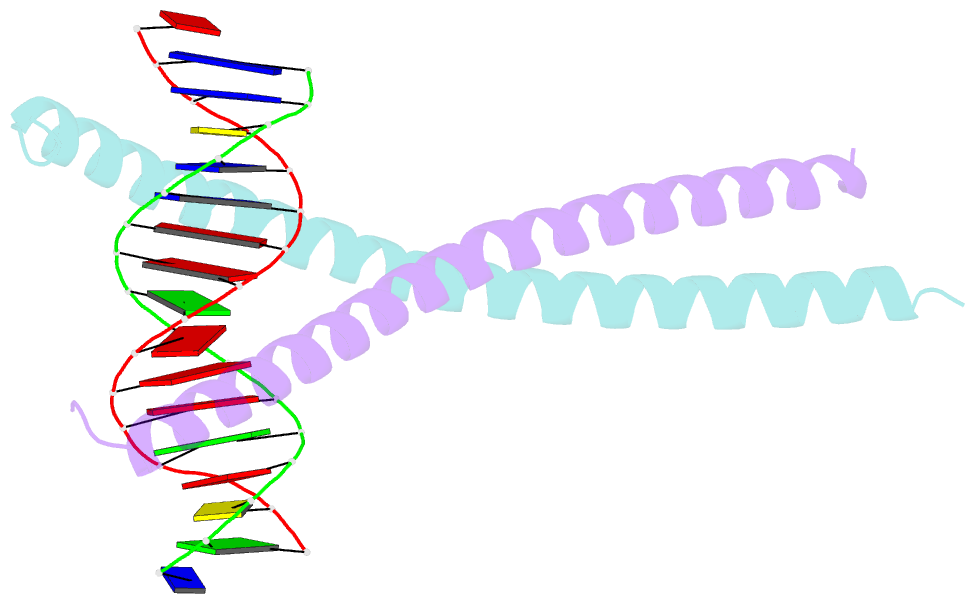
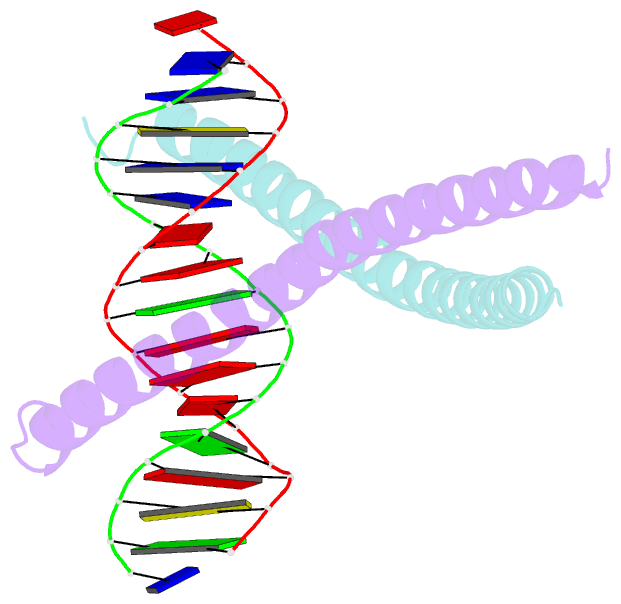
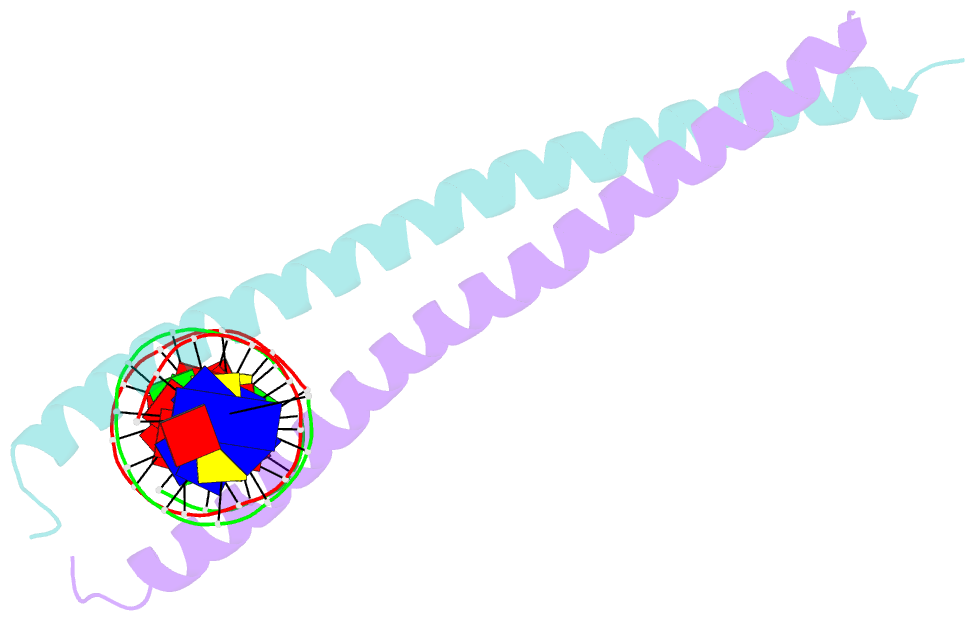
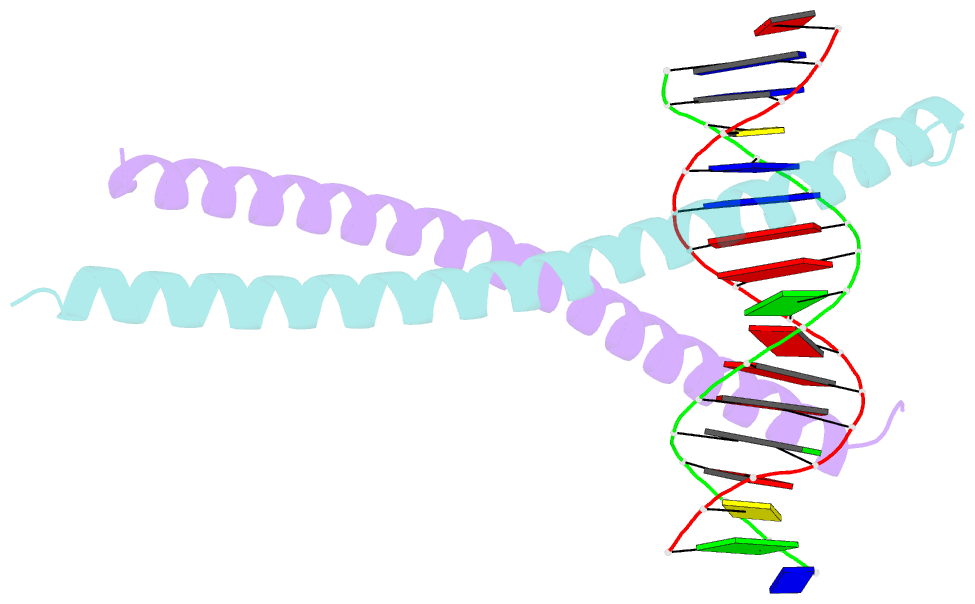
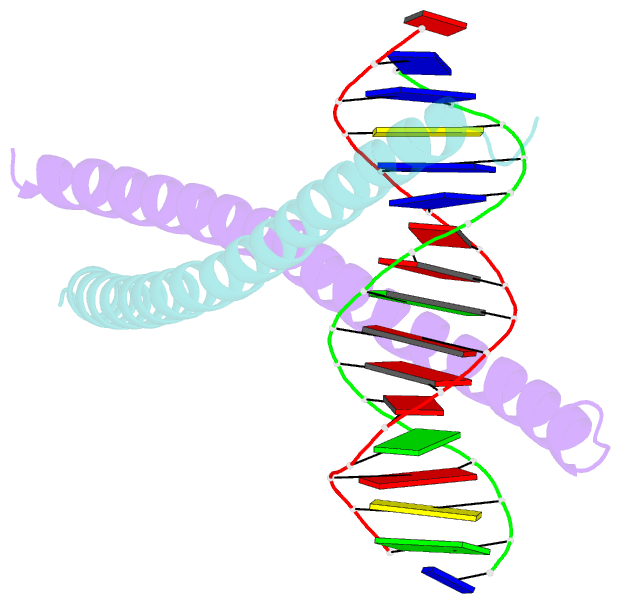
Summary information and primary citation
- PDB-id
- 7upz; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.487 Å)
- Summary
- Structural basis for cell type specific DNA binding of c-ebpbeta: the case of cell cycle inhibitor p15ink4b promoter
- Reference
- Lountos GT, Cherry S, Tropea JE, Wlodawer A, Miller M (2022): "Structural basis for cell type specific DNA binding of C/EBP beta : The case of cell cycle inhibitor p15INK4b promoter." J.Struct.Biol., 214, 107918. doi: 10.1016/j.jsb.2022.107918.
- Abstract
- C/EBPβ is a key regulator of numerous cellular processes, but it can also contribute to tumorigenesis and viral diseases. It binds to specific DNA sequences (C/EBP sites) and interacts with other transcription factors to control expression of multiple eukaryotic genes in a tissue and cell-type dependent manner. A body of evidence has established that cell-type-specific regulatory information is contained in the local DNA sequence of the binding motif. In human epithelial cells, C/EBPβ is an essential cofactor for TGFβ signaling in the case of Smad2/3/4 and FoxO-dependent induction of the cell cycle inhibitor, p15INK4b. In the TGFβ-responsive region 2 of the p15INK4b promoter, the Smad binding site is flanked by a C/EBP site, CTTAA•GAAAG, which differs from the canonical, palindromic ATTGC•GCAAT motif. The X-ray crystal structure of C/EBPβ bound to the p15INK4b promoter fragment shows how GCGC-to-AAGA substitution generates changes in the intermolecular interactions in the protein-DNA interface that enhances C/EBPβ binding specificity, limits possible epigenetic regulation of the promoter, and generates a DNA element with a unique pattern of methyl groups in the major groove. Significantly, CT/GA dinucleotides located at the 5'ends of the double stranded element maintain local narrowing of the DNA minor groove width that is necessary for DNA recognition. Our results suggest that C/EBPβ would accept all forms of modified cytosine in the context of the CpT site. This contrasts with the effect on the consensus motif, where C/EBPβ binding is modestly increased by cytosine methylation, but substantially decreased by hydroxymethylation.