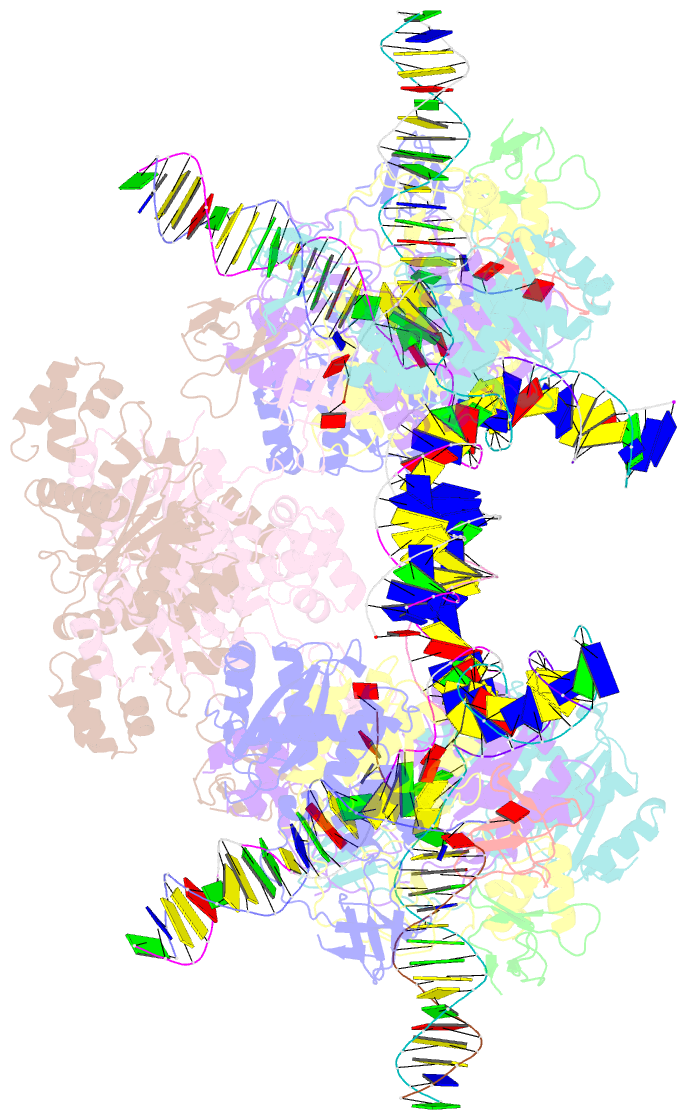
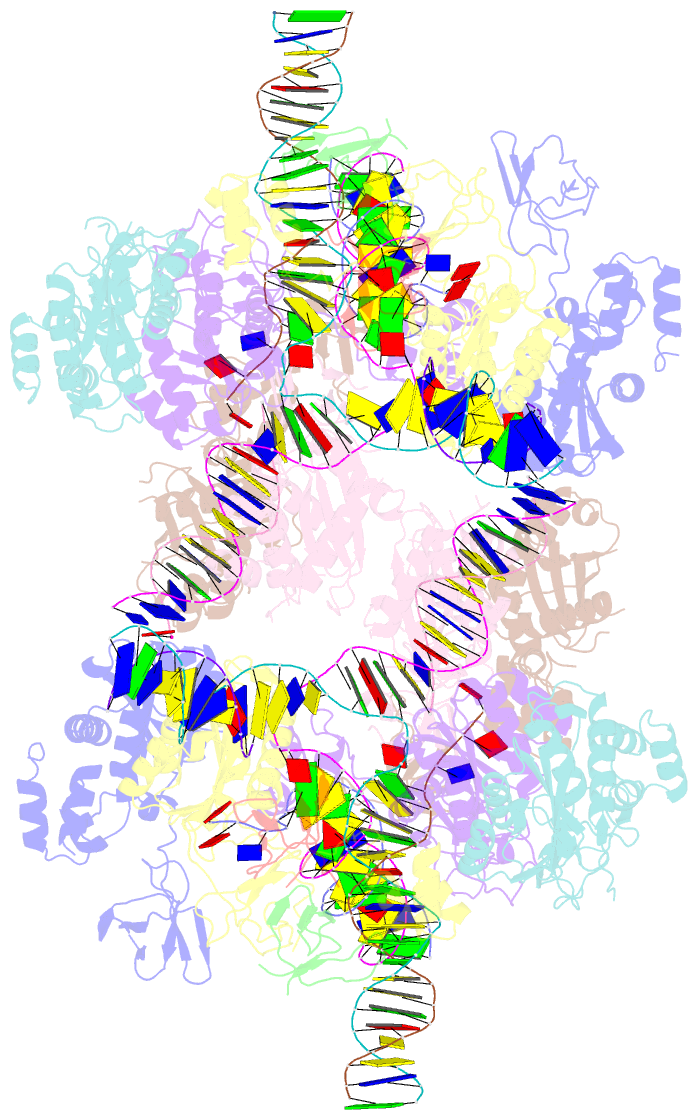
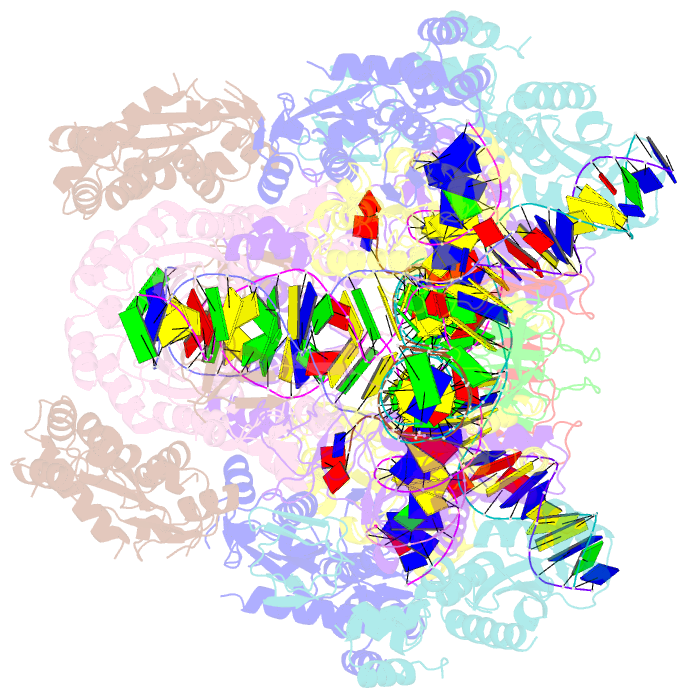
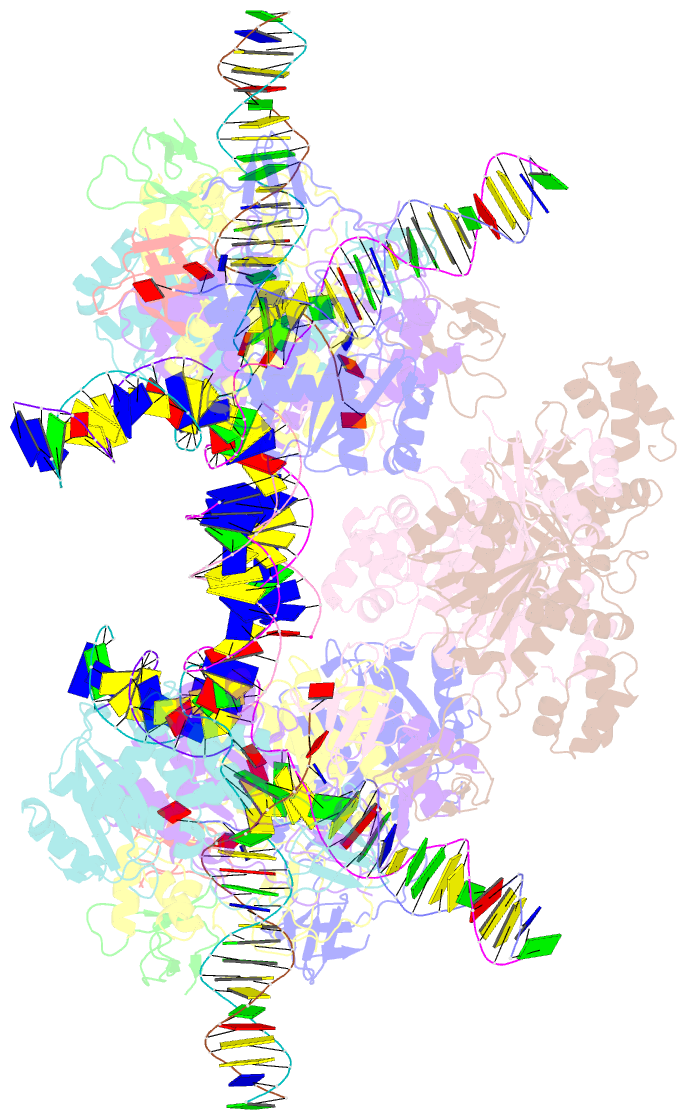
Summary information and primary citation
- PDB-id
- 7ut1; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- cryo-EM (3.8 Å)
- Summary
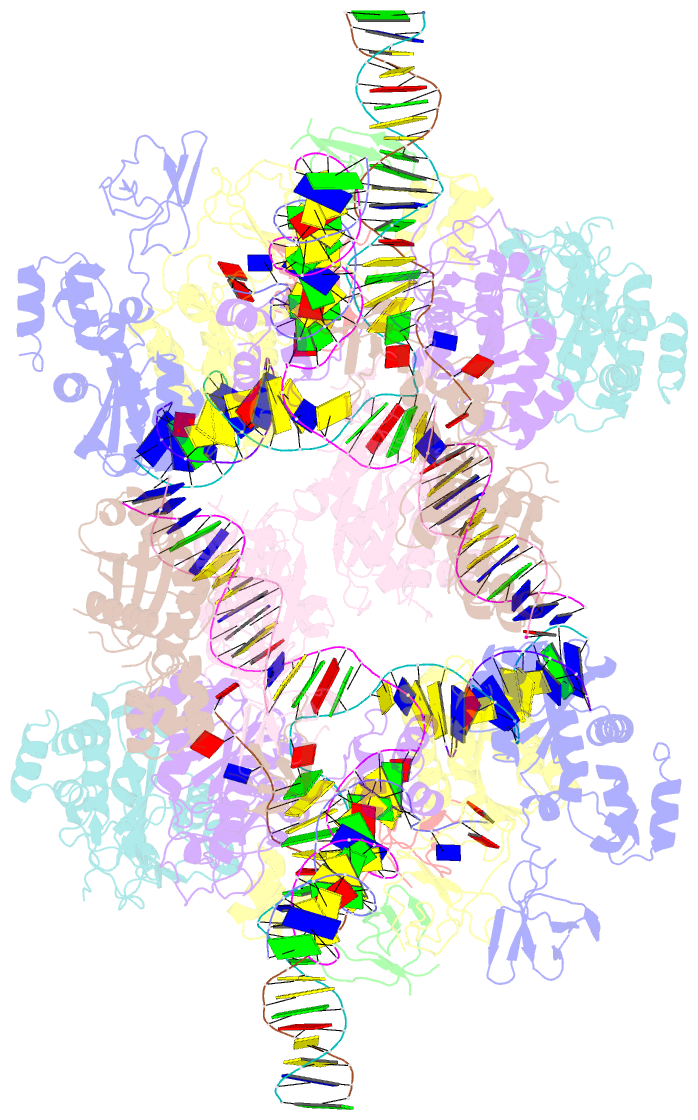
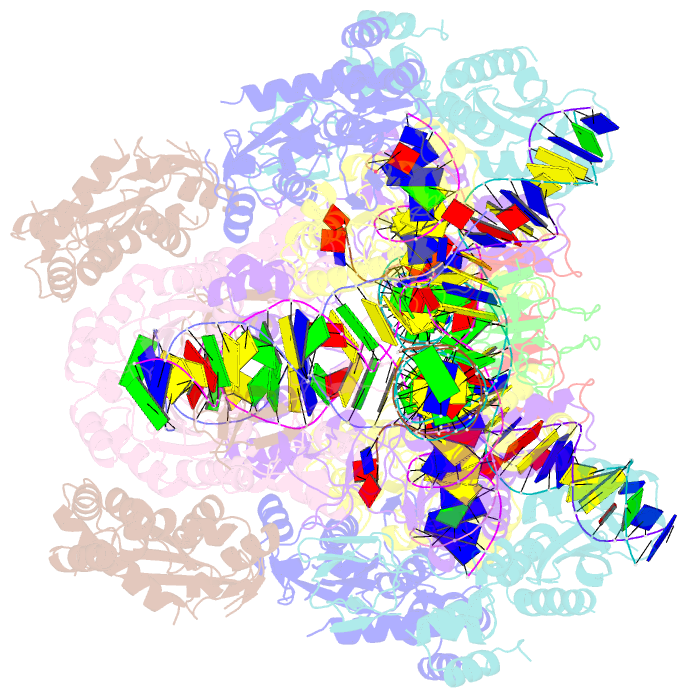
- Higher-order assembly of multiple mmtv strand transfer complex intasomes
- Reference
- Jozwik IK, Li W, Zhang DW, Wong D, Grawenhoff J, Ballandras-Colas A, Aiyer S, Cherepanov P, Engelman AN, Lyumkis D (2022): "B-to-A transition in target DNA during retroviral integration." Nucleic Acids Res., 50, 8898-8918. doi: 10.1093/nar/gkac644.
- Abstract
- Integration into host target DNA (tDNA), a hallmark of retroviral replication, is mediated by the intasome, a multimer of integrase (IN) assembled on viral DNA (vDNA) ends. To ascertain aspects of tDNA recognition during integration, we have solved the 3.5 Å resolution cryo-EM structure of the mouse mammary tumor virus (MMTV) strand transfer complex (STC) intasome. The tDNA adopts an A-like conformation in the region encompassing the sites of vDNA joining, which exposes the sugar-phosphate backbone for IN-mediated strand transfer. Examination of existing retroviral STC structures revealed conservation of A-form tDNA in the analogous regions of these complexes. Furthermore, analyses of sequence preferences in genomic integration sites selectively targeted by six different retroviruses highlighted consistent propensity for A-philic sequences at the sites of vDNA joining. Our structure additionally revealed several novel MMTV IN-DNA interactions, as well as contacts seen in prior STC structures, including conserved Pro125 and Tyr149 residues interacting with tDNA. In infected cells, Pro125 substitutions impacted the global pattern of MMTV integration without significantly altering local base sequence preferences at vDNA insertion sites. Collectively, these data advance our understanding of retroviral intasome structure and function, as well as factors that influence patterns of vDNA integration in genomic DNA.