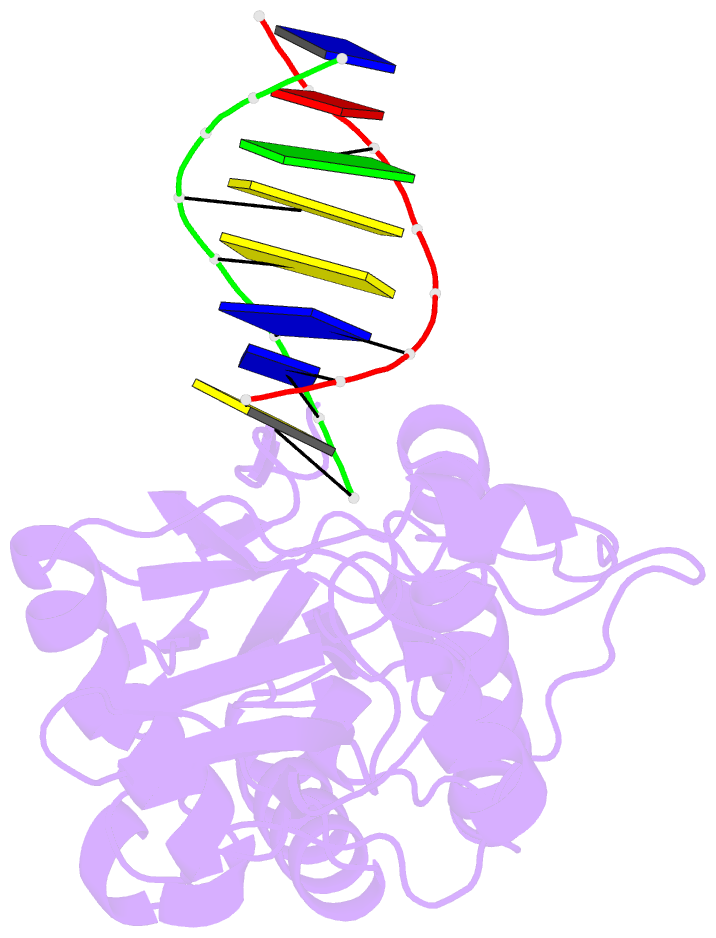
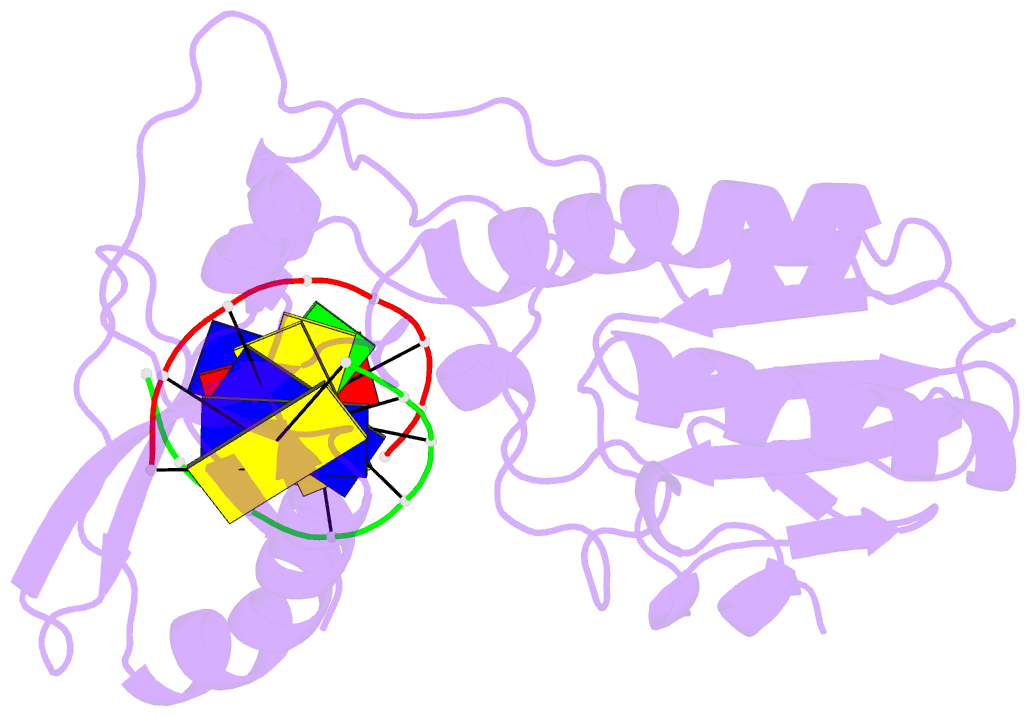
Summary information and primary citation
- PDB-id
- 7uyp; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (1.5 Å)
- Summary
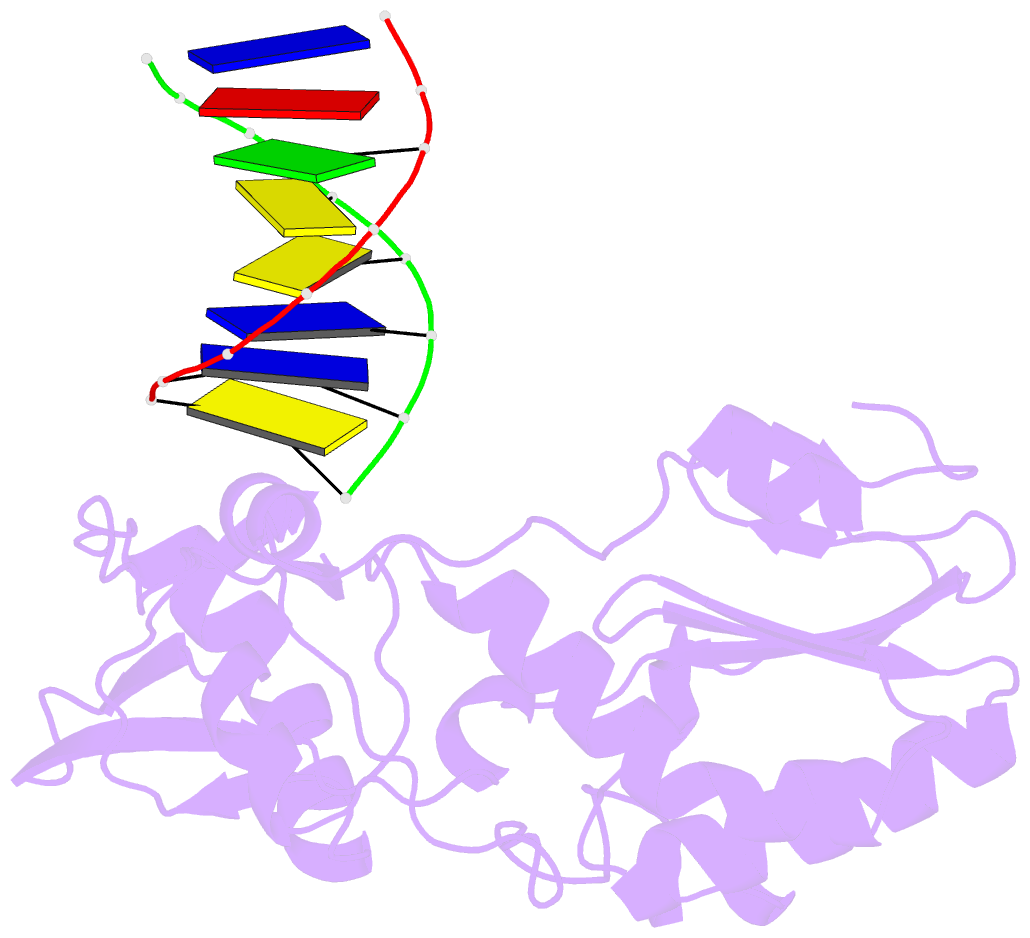
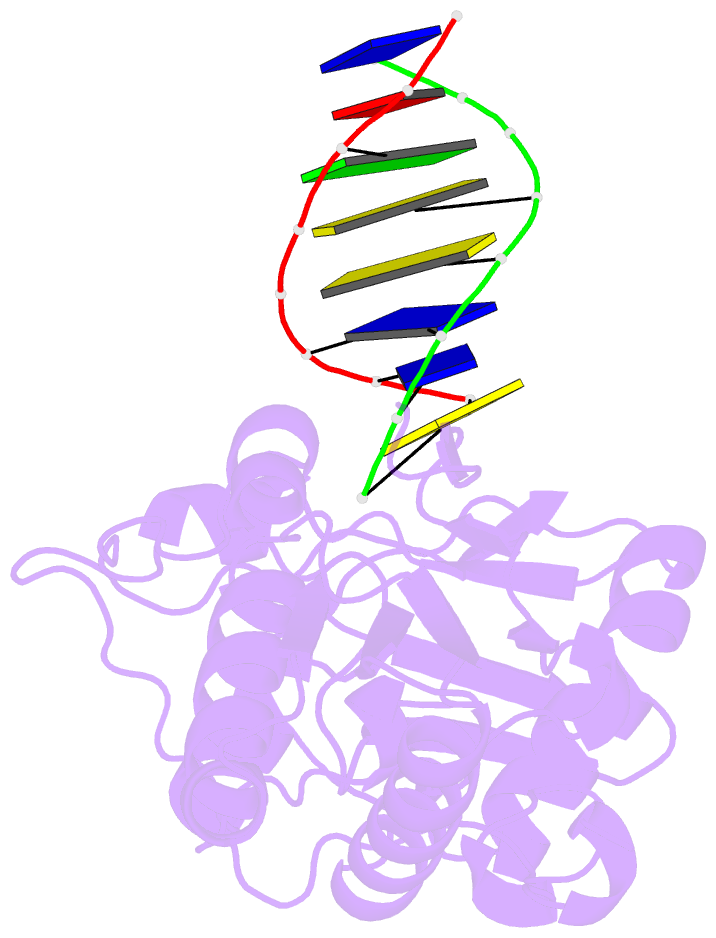
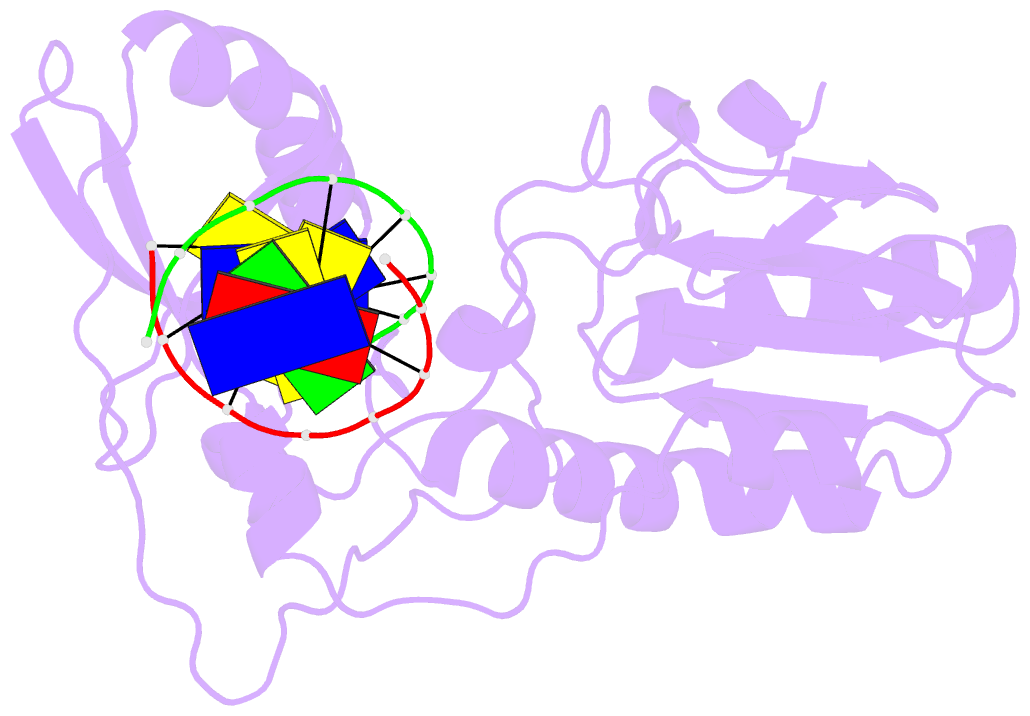
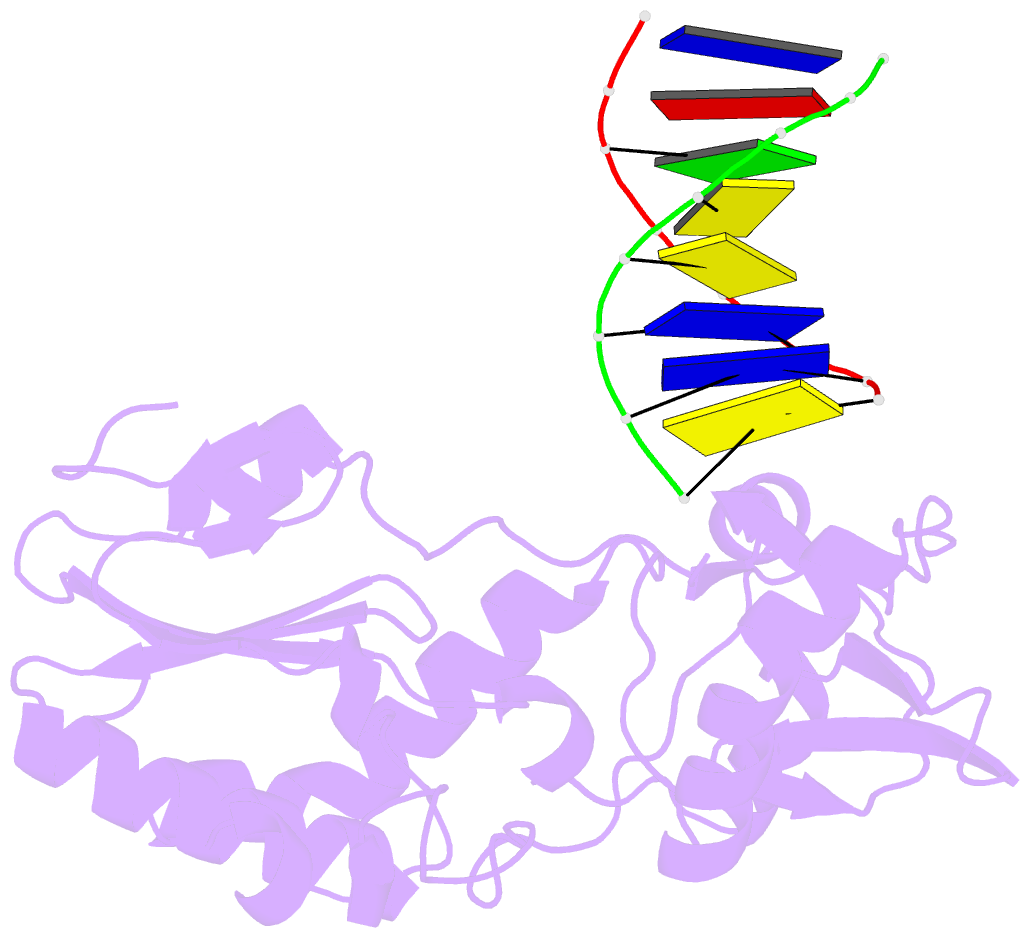
- Crystal structure of b-form alien DNA 5'-cttzzpbsbszppaag in a host-guest complex with the n-terminal fragment of moloney murine leukemia virus reverse transcriptase
- Reference
- Shukla MS, Hoshika S, Benner SA, Georgiadis MM (2023): "Crystal structures of 'ALternative Isoinformational ENgineered' DNA in B-form." Philos.Trans.R.Soc.Lond.B Biol.Sci., 378, 20220028. doi: 10.1098/rstb.2022.0028.
- Abstract
- The first structural model of duplex DNA reported in 1953 by Watson & Crick presented the double helix in B-form, the form that genomic DNA exists in much of the time. Thus, artificial DNA seeking to mimic the properties of natural DNA should also be able to adopt B-form. Using a host-guest system in which Moloney murine leukemia virus reverse transcriptase serves as the host and DNA as the guests, we determined high-resolution crystal structures of three complexes including 5'-CTT