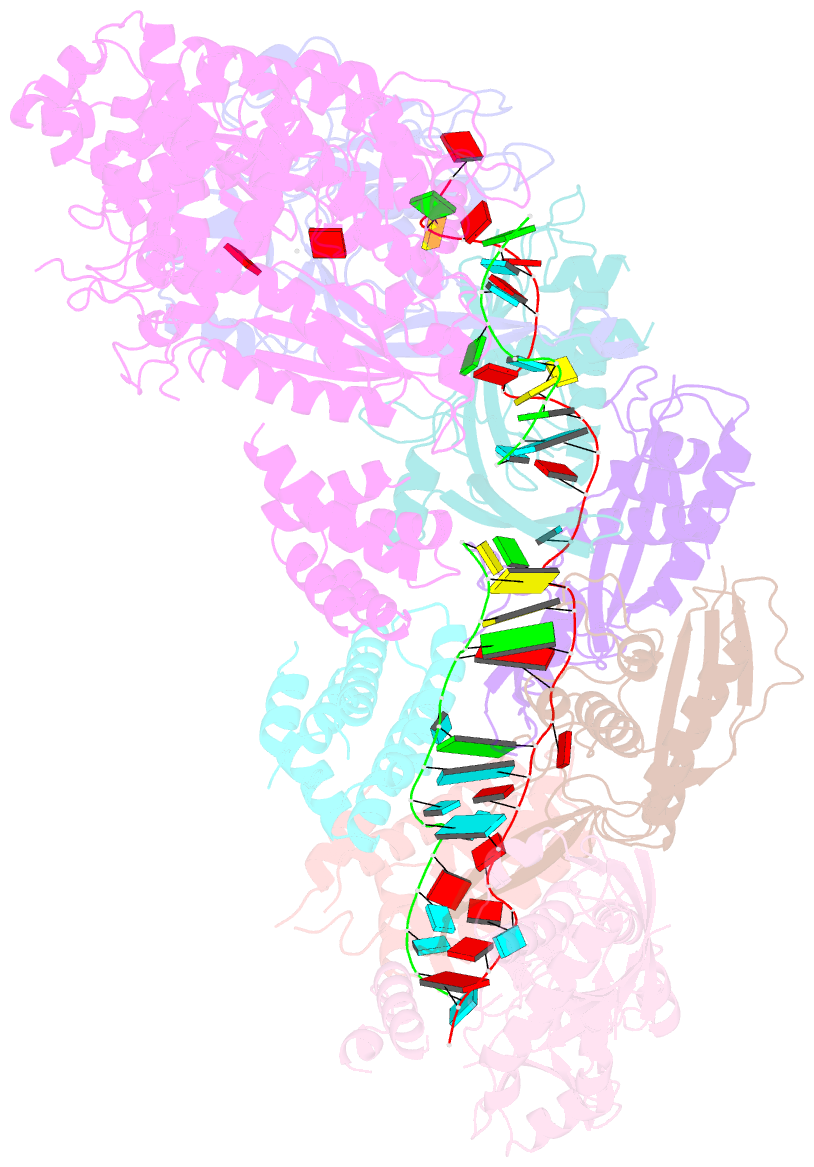
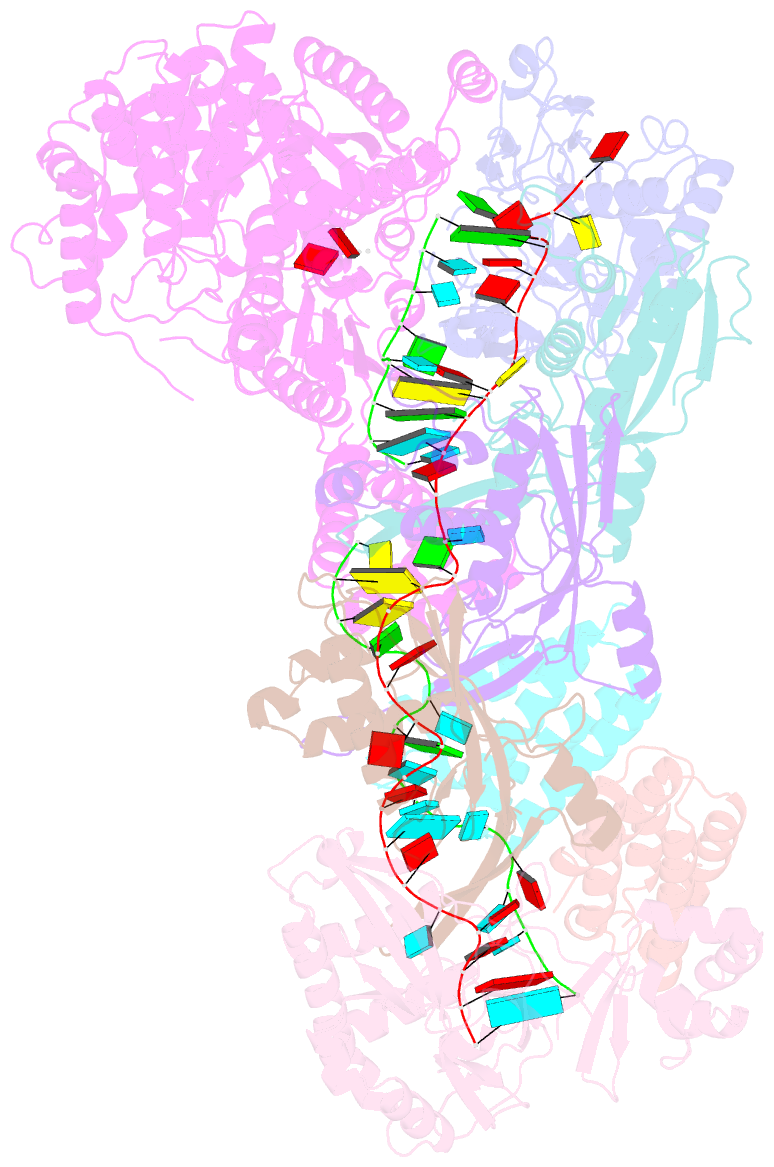
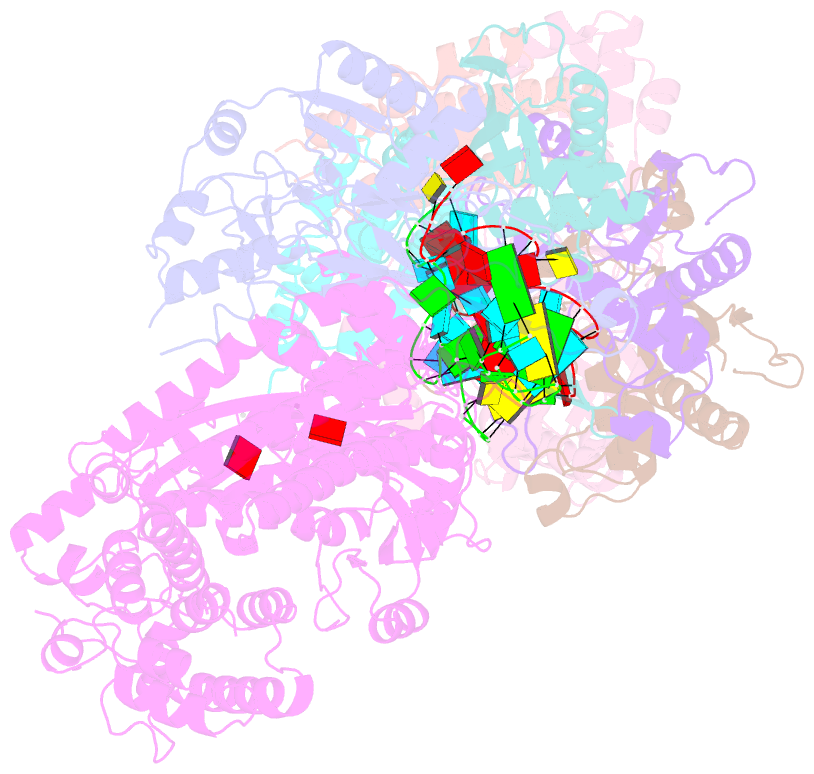
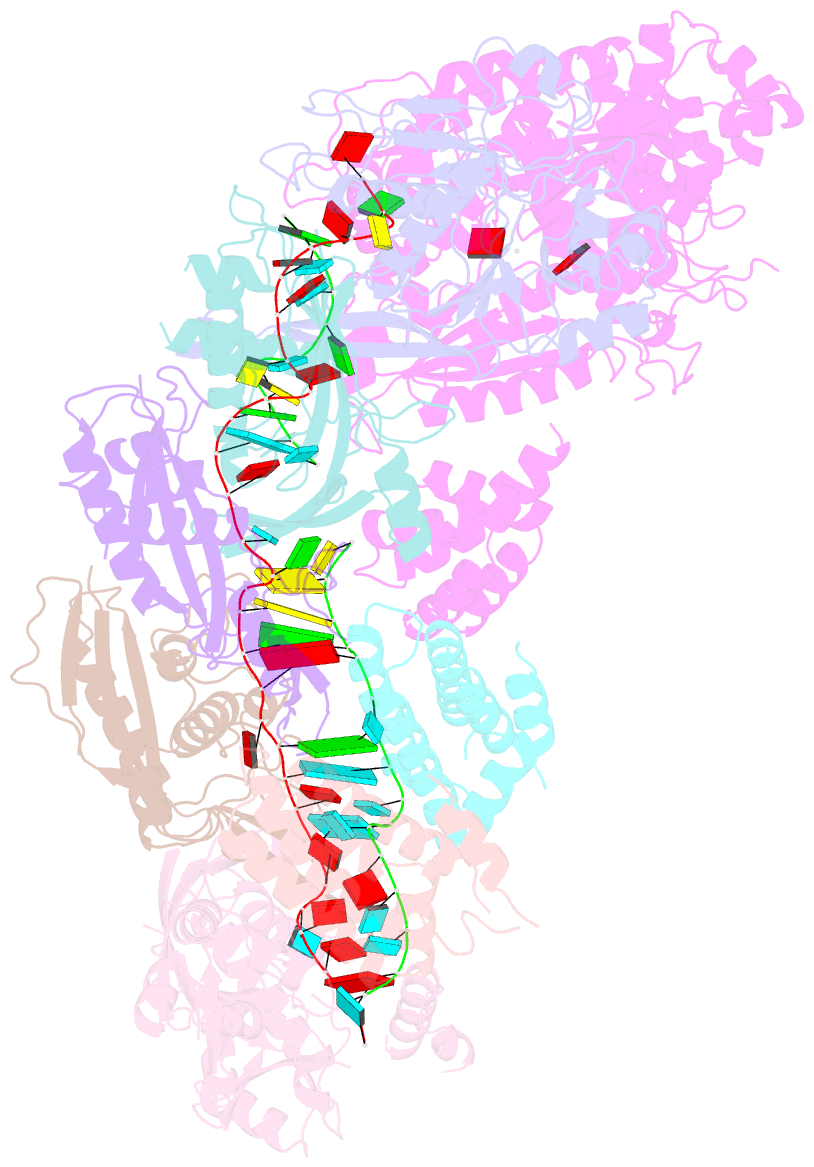
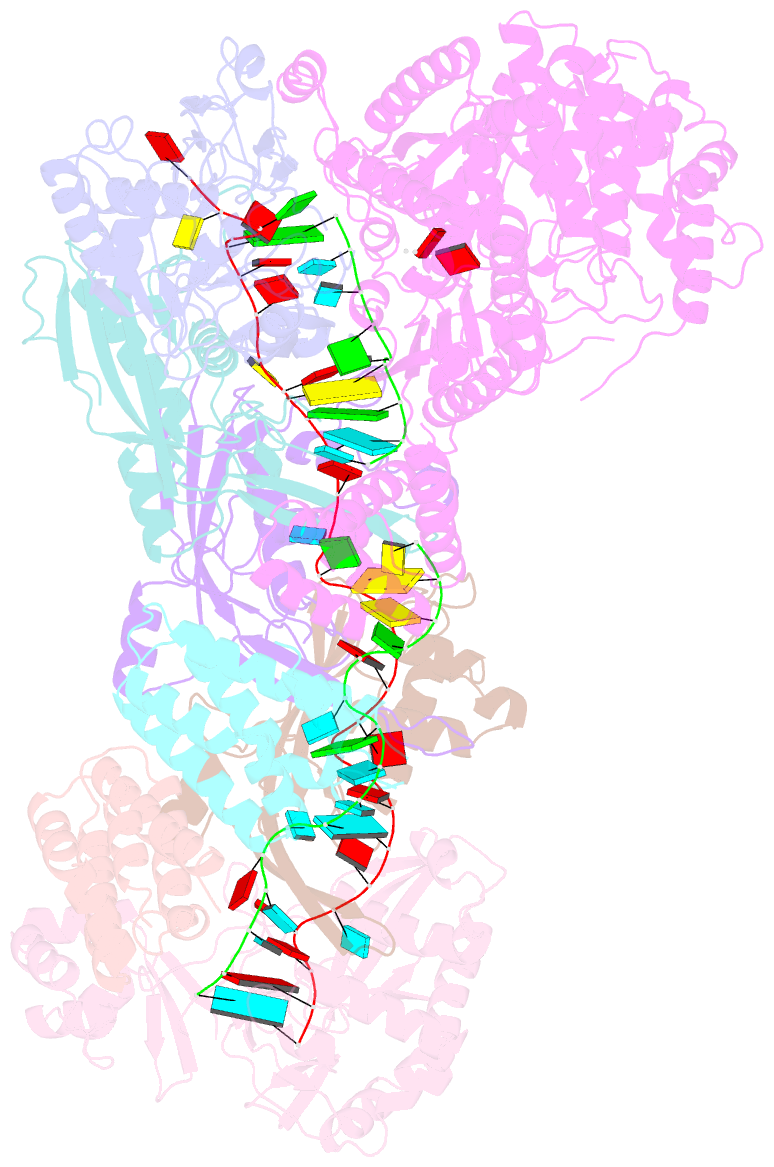
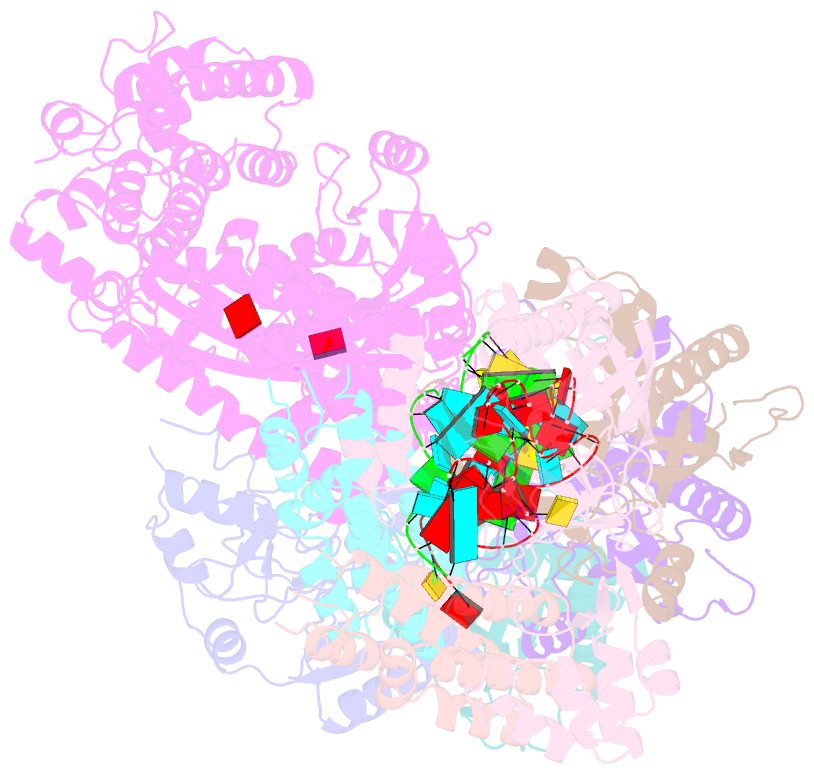
Summary information and primary citation
- PDB-id
- 7v01; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-RNA
- Method
- cryo-EM (3.67 Å)
- Summary
- Staphylococcus epidermidis rp62a crispr short effector complex with self RNA target and atp
- Reference
- Smith EM, Ferrell S, Tokars VL, Mondragon A (2022): "Structures of an active type III-A CRISPR effector complex." Structure, 30, 1109-1128.e6. doi: 10.1016/j.str.2022.05.013.
- Abstract
- Clustered regularly interspaced short palindromic repeats (CRISPR) and their CRISPR-associated proteins (Cas) provide many prokaryotes with an adaptive immune system against invading genetic material. Type III CRISPR systems are unique in that they can degrade both RNA and DNA. In response to invading nucleic acids, they produce cyclic oligoadenylates that act as secondary messengers, activating cellular nucleases that aid in the immune response. Here, we present seven single-particle cryo-EM structures of the type III-A Staphylococcus epidermidis CRISPR effector complex. The structures reveal the intact S. epidermidis effector complex in an apo, ATP-bound, cognate target RNA-bound, and non-cognate target RNA-bound states and illustrate how the effector complex binds and presents crRNA. The complexes bound to target RNA capture the type III-A effector complex in a post-RNA cleavage state. The ATP-bound structures give details about how ATP binds to Cas10 to facilitate cyclic oligoadenylate production.